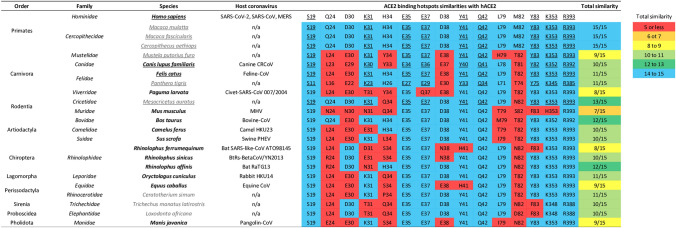
Figure 2.
Comparisons at key ACE2 sites between human and a sample of mammals belong to ten orders. Species that have been experimentally identified as susceptible to SARS-CoV-2 infection are underlined. Species that can be infected by their own host-specific coronaviruses are in black bold, and other species are in grey. Highlighted blue and red are matching and mis-matching amino acid residues, respectively, between mammalian ACE2 and human ACE2 at key SARS-CoV-2 S protein binding sites. Underlined amino acids are conserved among species known to be infected by SARS-CoV-2. Total similarity designates the total number of matching amino acid residues with respect to human ACE2. The total similarity score for mammalian ACE2 in blue highlights perfect site similarities (14–15 matching sites), in green highlights high similarity (12–13 matching sites), in light green highlights medium–high similarity (10–11 matching sites), in yellow highlights medium similarity (8–9 matching sites), in orange highlights medium–low similarity (6–7 matching sites), and in red highlights low similarity (5 or less matching sites). A comparison of key ACE2 sites involved in SARS-CoV-2 binding between human and 131 mammals is shown in Supplementary Figure S1.