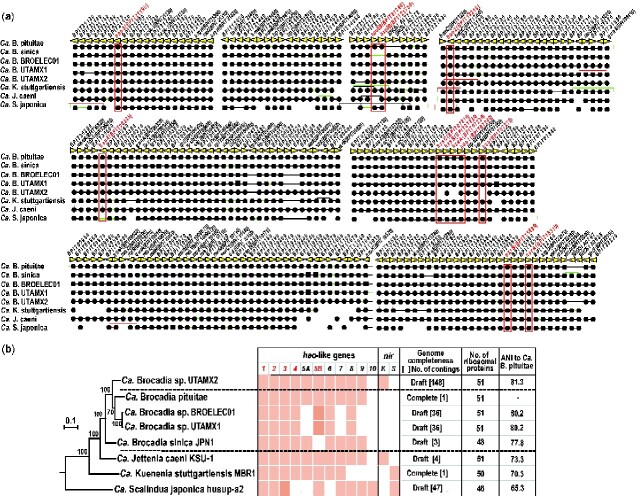
Figure 5.
Phylogenetic relationships and core gene set among anammox bacteria. (a) Phylogenetic tree based on amino acid sequences of the concatenated ribosomal proteins of anammox bacteria and retention patterns of hao-like and nitrite reductase nirS and nirK, genes. Two complete genomes Ca. B. pituitae and Ca. K. stuttgartiensis, and six nearly completed draft genomes were used for this study. (b) Core gene alignment of anammox bacteria. Rows and columns represent genomes and OGs, respectively. Black lines represent direct adjacency, green lines represent non-adjacent neighbourhoods indicating the existence of insertions, and red lines represent inversions. Closed circles and squares indicate single and paralogous genes, respectively. Dashed lines show the connection to the divided gene. Red characters show genes associated with anammox reactions. ANI: average nucleotide identity. The ANI values show identity % of Ca. B. pituitae genome to other anammox bacterial with nearly complete genome.