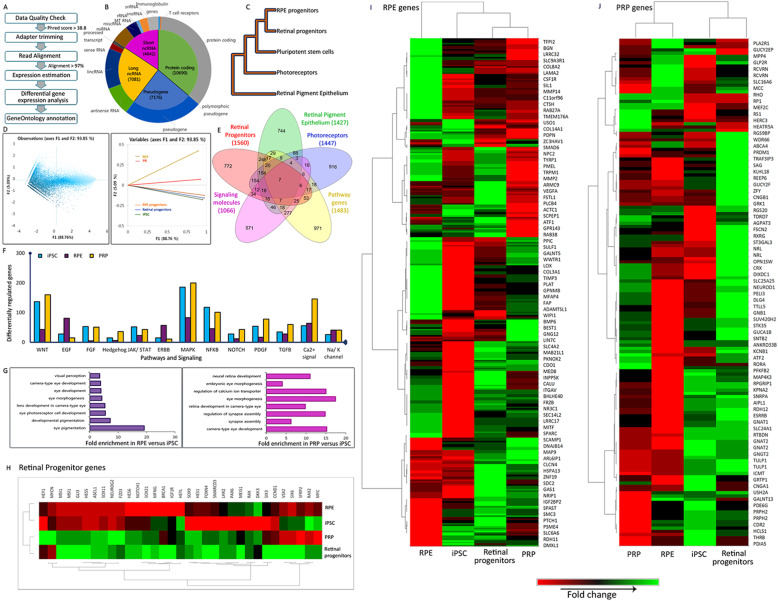
Fig. 5.
RNAseq data analysis and interpretation. a Flow chart of RNAseq workflow and bioinformatic analysis. b Pie chart showing the distribution of genes picked up by samples, categories further explained by doughnut chart. c Hierarchical clustering of samples using the UPGMA method. d Principal component analysis showing the relation between active variables. e Five sample Venn diagrams of differentially expressed genes from retinal progenitors, RPE, and PRP; significantly expressed pathway genes and signaling molecules. f Column graph showing the regulation of various pathways in iPSC, RPE, and PRP samples. g Gene Ontology and fold enrichment analysis for RPE and PRP. h–j Heat map shows clustering and differential expression of key genes from RNAseq data across iPSC, day 20 retinal progenitors, and day 70 RPE and PRP. h Retinal Progenitors. i RPE signature genes. j Rod and cone-specific PRP genes. Heat map colors: red to green through black