FIGURE 6.
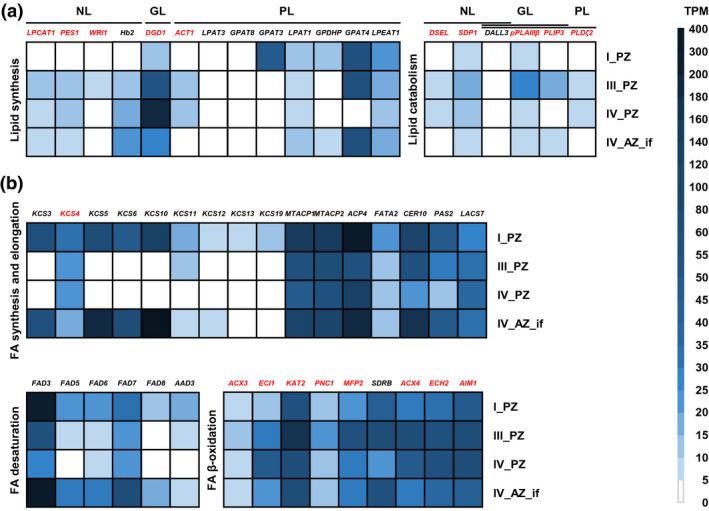
Gene expression profiles of 50 individual genes out of the NL, GL, PL, and FA‐related GO categories. Represented are RNA‐seq data (TPM, transcripts per million) of A. alpina perpetual flowering 1‐1 (pep1‐1) mutant lateral stem internodes of perennial (PZ) and annual (AZ) zones of RNA‐seq at stages I_PZ, III_PZ, IV_PZ, and IV_AZ_if (see Figure 5a). (a) NL, GL, and PL genes involved in lipid synthesis and catabolism: LPCAT1, LYSOPHOSPHATIDYLCHOLINE ACYLTRANSFERASE 1, PES1, PHYTYL ESTER SYNTHASE 1, WRI1, WRINKLED 1, Hb2, HAEMOGLOBIN 2, DGD1, DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1, ACT1, ACYLTRANSFERASE 1, LPAT3, LYSOPHOSPHATIDYL ACYLTRANSFERASE 3, GPAT8, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 8, GPAT3, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 3, LPAT1, LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE 1, GPDHP, GLYCEROL‐3‐PHOSPHATE DEHYDROGENASE PLASTIDIC, GPAT4, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 4, LPEAT1, LYSOPHOSPHATIDYLETHANOLAMINE ACYLTRANSFERASE 1, DSEL, DAD1‐LIKE SEEDLING ESTABLISHMENT‐RELATED LIPASE, SDP1, SUGAR‐DEPENDENT1, DALL3, DAD1‐LIKE LIPASE 3, pPLAIIIβ, PATATIN‐RELATED PHOSPHOLIPASE A IIIβ, PLIP3, PLASTID LIPASE 3, PLDζ2, PHOSPHOLIPASE D ζ 2; (b) Genes involved in FA synthesis, elongation, desaturation, and β‐oxidation: KCS3, KCS4, KCS5, KCS6, KCS10, KCS11, KCS12, KCS13, KCS19, KETOACYL‐COA SYNTHASEs, MTACP1, MITOCHONDRIAL ACYL CARRIER PROTEIN 1, MTACP2, MITOCHONDRIAL ACYL CARRIER PROTEIN 2, ACP4, ACYL CARRIER PROTEIN 4, FATA2, OLEOYL‐ACYL CARRIER PROTEIN THIOESTERASE 2, CER10, ECERIFERUM 10, PAS2, PASTICCINO 2, LACS7, LONG‐CHAIN ACYL‐COA SYNTHETASE 7, FAD3, FAD5, FAD6, FAD7, FAD8, FATTY ACID DESATURASEs, AAD3, ACYL‐ACYL CARRIER PROTEIN DESATURASE 3, ACX3, ACYL‐COA OXIDASE 3, ECI1, Δ3, Δ2‐ENOYL COA ISOMERASE 1, KAT2, 3‐KETOACYL‐COA THIOLASE 2, PNC1, PEROXISOMAL ADENINE NUCLEOTIDE CARRIER 1, MFP2, MULTIFUNCTIONAL PROTEIN 2, SDRB, SHORT‐CHAIN DEHYDROGENASE‐REDUCTASE B, ACX4, ACYL‐COA OXIDASE 4, ECH2, ENOYL‐COA HYDRATASE 2, AIM1, ABNORMAL INFLORESCENCE MERISTEM 1. Candidate genes differentially regulated between PZ and AZ are represented in red. Data are represented as mean (n = 3)
