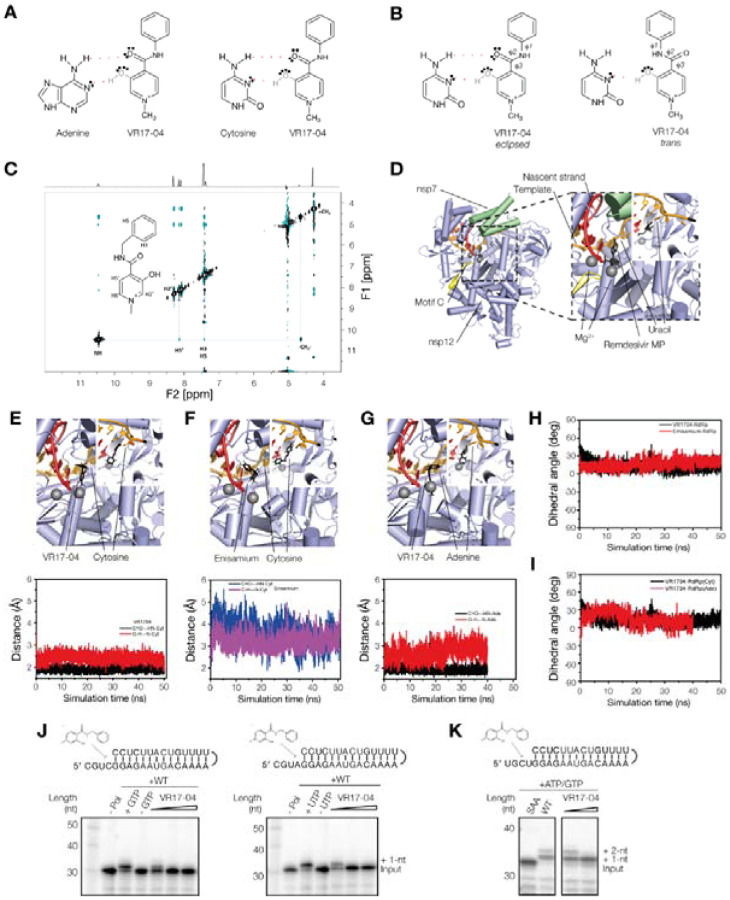
Figure 2. Enisamium metabolite VR17–04.
(A) Schematic of putative hydrogen bond formation between cytosine and adenine bases with VR17–04. (B) Schematic of the trans and eclipsed conformations of VR17–04. (C) 2D-NOESY and 1H proton (above) spectra of VR17–04 acquired at 277 K in water. The NOE correlation between the HN and H5’ proton is highlighted with a dashed line. (D) Structure of the SARS-CoV-2 nsp12/7/8 complex bound to RNA and remdesivir monophosphate. Rendering based on PDB 7bv2. (E) Model (top) and MD simulation (bottom) of VR17–04 binding to cytosine in nsp12 active site. (F) Model (top) and MD simulation (bottom) of enisamium binding to cytosine in nsp12 active site. (G) Model (top) and MD simulation (bottom) of VR17–04 binding to adenine in nsp12 active site. (H) MD simulation of dihedral angle of VR17–04 or enisamium binding to cytosine in nsp12 active site. (I) MD simulation of dihedral angle of VR17–04 binding to cytosine or adenine in nsp12 active site (J) Effect of VR17–04 on SARS-CoV-2 nsp12/7/8 activity on two different hairpin templates in the presence of GTP (left) or UTP (right). In the presence of wildtype nsp12/7/8 and GTP or UTP, the radiolabelled primer was extended by 1 nt. (K) Effect of VR17–04 on SARS-CoV-2 nsp12/7/8 primer extension activity. A mutant containing a double amino acid substitution in the nsp12 active site (SDD=>SAA) was used as negative control. ATP and GTP were added to the reaction to allow extension of the template by 2 nt.