FIGURE 2.
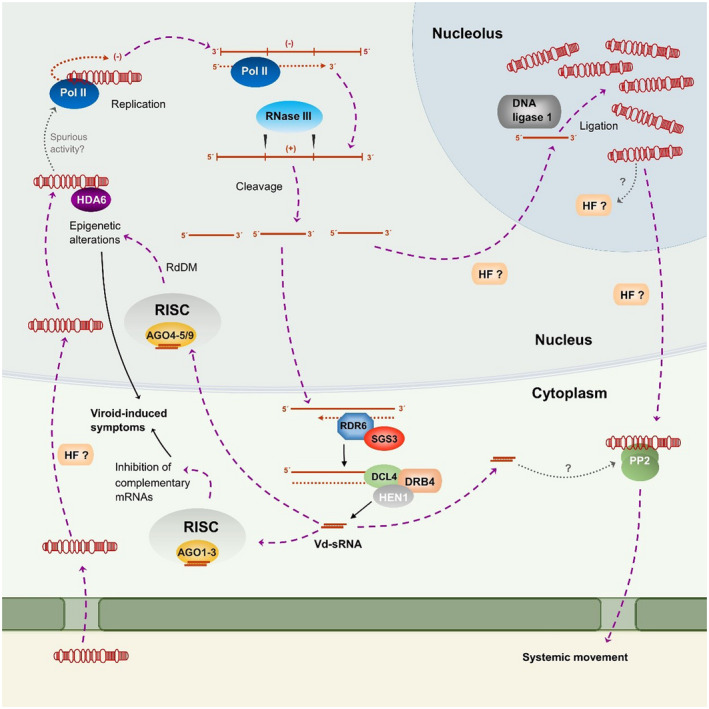
Proposed model of HSVd cell cycle and interactions with host factors. HSVd genomic RNA enters the cell through the plasmodesmata and it is trafficked to the nucleus, probably assisted by unidentified host factors. In the nucleus, HSVd subverts the activity of histone deacetylase 6 (HDA6) and causes epigenetic alterations. The subversion of HDA6 may favour a spurious recognition of the genomic HSVd RNA by the DNA‐dependent‐RNA polymerase II (Pol II). HSVd replication follows an asymmetric rolling circle in which Pol II transcribes HSVd RNAs. Oligomeric RNAs are cleaved by a host enzyme with RNase III activity and the resultant monomers are circularized by DNA ligase I, probably in the nucleolus, in which it is assumed that mature HSVd molecules accumulate. Other HSVd replication intermediates are recognized as aberrant transcripts and enter the trans‐acting small interfering RNA (tasiRNA) biogenesis pathway. Therefore, these RNAs are trafficked to the cytoplasm, transcribed by RDR6, and processed by DCL4 into viroid‐derived small RNAs (vd‐sRNAs) that are loaded into different AGO proteins. Vd‐sRNAs of 21–22 nucleotides are loaded in AGO 1‐3 in the RISC complex and direct the inhibition of host mRNA transcripts in the cytoplasm. Moreover, vd‐sRNAs of 24 nucleotides are primarily responsible for RNA‐directed DNA methylation (RdDM) in the nucleus. Thus, HSVd‐sRNAs cause transcriptional (nucleus) and posttranscriptional (cytoplasm) alterations that might account for the viroid‐induced symptoms. Mature HSVd genomic RNAs are exported to the phloem by forming a ribonucleoprotein complex with the phloem protein 2 (PP2). Additionally, the systemic movement of HSVd‐sRNA may also be possible due to the interaction with PP2
