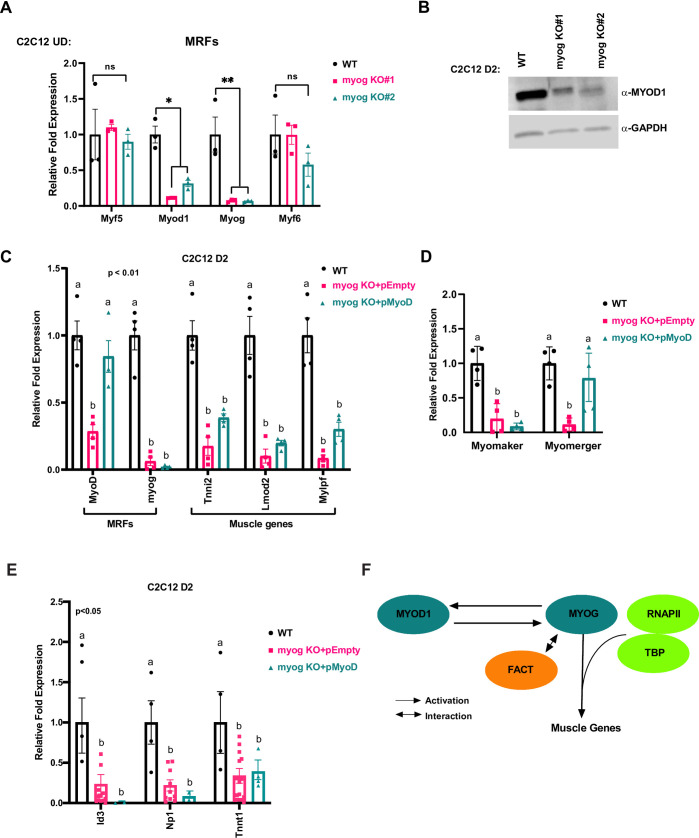
Fig 6. MYOG regulates MYOD1 in C2C12 cells and is required for muscle gene activation.
A. C2C12 clones with the Myog deletion were grown in proliferating condition (U.D.) and were harvested for total RNA and assayed for the expression of Myf5, Myod1, Myog, and Myf6 by qRT-PCR. Standard errors (S.E.) from the mean (Mean ± S.E.) represents the error bars. (Student t.test; ns represents ‘not significant’, *p<0.05 and **p<0.01, n = 3 biological replicates). B. Cells as in A were differentiated for two days (D2) and were assayed for protein expression of MYOD1 by western blot. GAPDH was used as a loading control. C-E. Cells as in A were stably transfected with MYOD1 expression construct (pMyoD) or empty plasmid (pEmpty). The cells were then selected and assayed for the mRNA expression of Myod1, Myog, Tnni2, Lmod2, Mylpf (C), Myomaker, Myomerger (D) and the MYOD1 target genes, Id3, Np1 and Tnnt1 (E) by qRT-PCR. Standard errors (S.E.) from the mean (Mean ± S.E.) represents the error bars. (ANOVA test followed by Tukey's multiple comparisons test for each gene; Samples not sharing an alphabet within a gene group are statistically significant, p<0.01 and n = 3–4 biological replicates). F. A schematic model representing MYOG dependent regulation of muscle genes transcription as well as MYOD1, an upstream regulator of Myog expression during skeletal muscle differentiation.