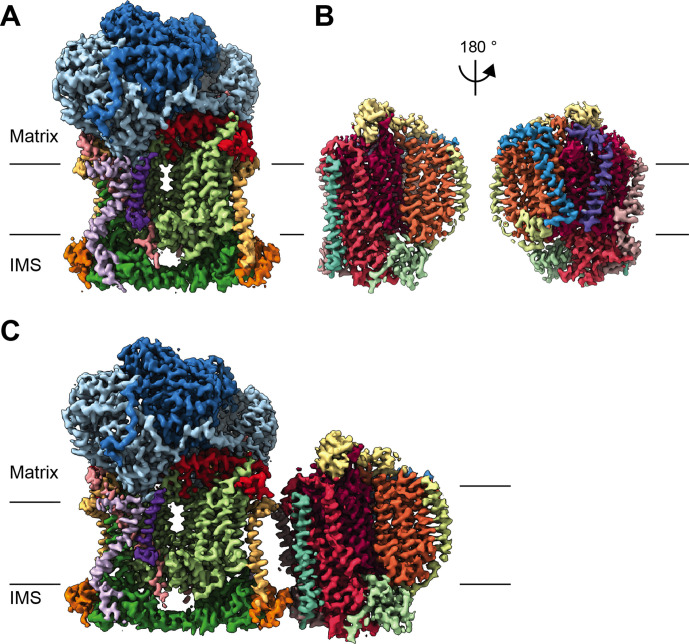
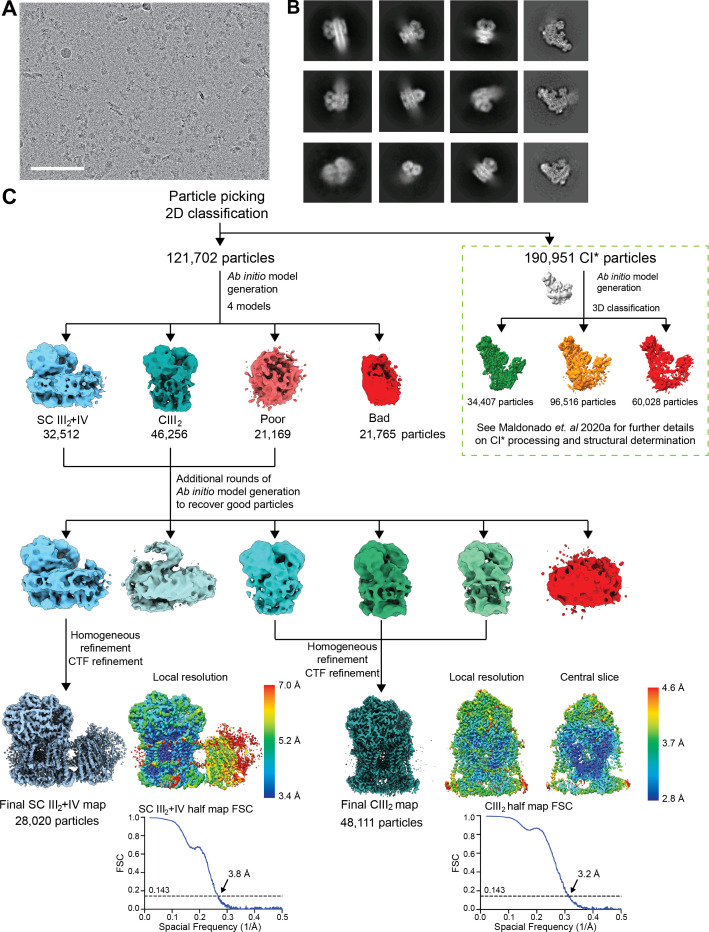
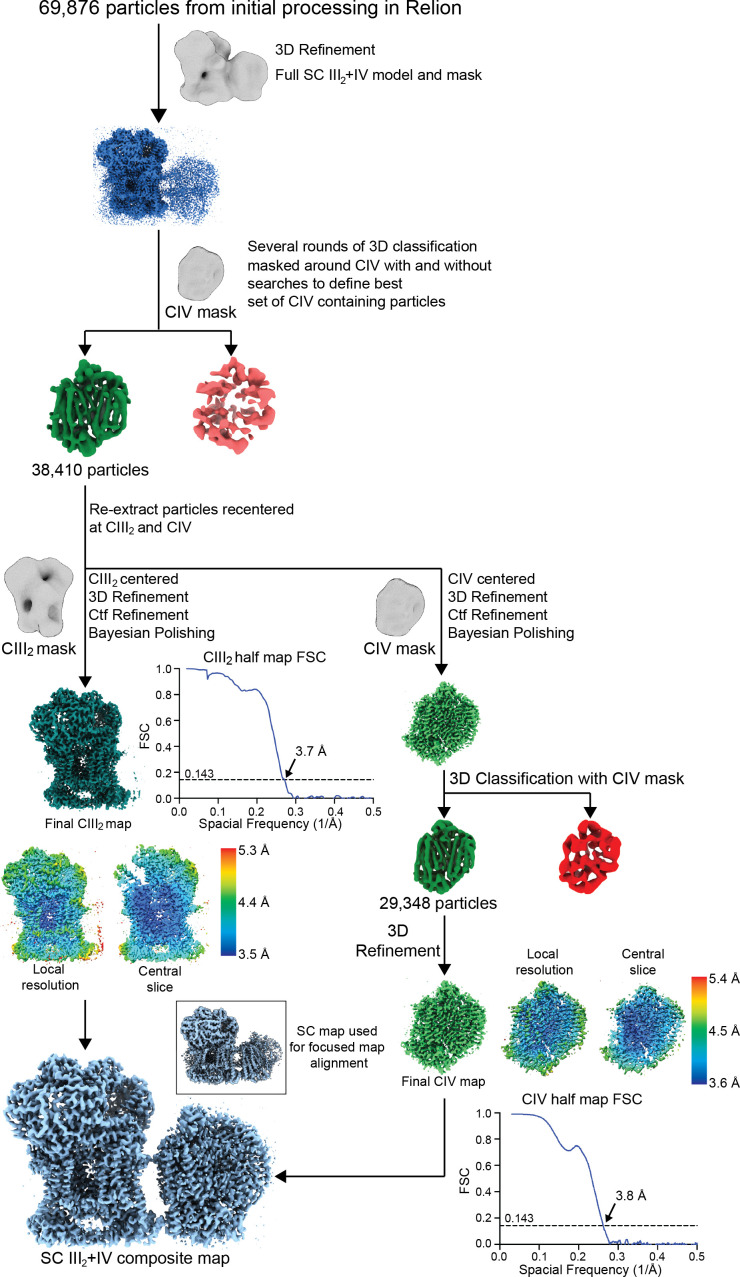
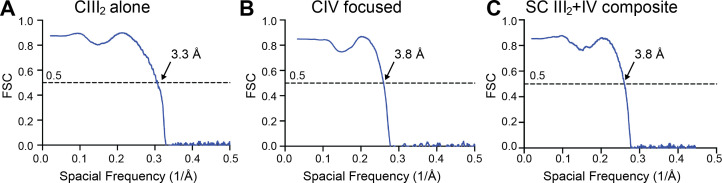
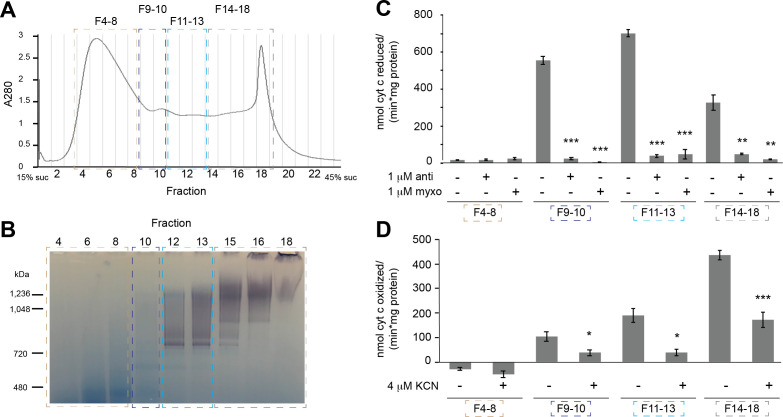
(A) Digitonin-extracted, amphipol-stabilized V. radiata mitochondrial membrane sample was separated by a 15–45% (w:v) linear sucrose (suc) gradient and fractionated. Relevant fractions were pooled and concentrated as indicated by dashed boxes and labels (fractions 4–8, 9–10, 11–13, 14–18) after the in-gel activity assay in (B). (B) Select fractions of the sucrose gradient fractions from (A) were run on a BN-PAGE gel and subjected to in-gel NADH-dehydrogenase activity assay. Note that not all fractions were loaded on the gel, and that the peak on fraction 18 corresponds to aggregation, likely due to the use of old mitochondrial samples due to COVID-19-related research restrictions. (C) The activity of the pooled samples from (A) was tested with a spectroscopic activity assay from reduced-decylubiquinone to cytochrome c, in the presence or absence of 2 µM antimycin (anti) or myxothiazol (myxo). Three to five independent repeat measurements were done for each sample. The background-corrected average of the repeats is shown, together with the standard error from the mean (S.E.M., error bars). Significance (**, p<0.01; ***, p<0.001) was tested with two-tailed t-tests for each inhibitor-exposed sample with respect to the control. p-values from left to right: 0.97, 0.32, 8.0 × 10−5, 1.5 × 10−4, 4.7 × 10−6, 2.8 × 10−5, 0.0064, 0.0051. (D) The activity of the pooled samples from (A) was tested with a spectroscopic activity assay to follow the oxidation of reduced cytochrome c, in the presence or absence of 4 µM potassium cyanide (KCN). Three to four independent repeat measurements were done for each sample. The background-corrected average of the repeats is shown, together with the standard error from the mean (S.E.M., error bars). Significance (*, p<0.05; ***, p<0.001) was tested with two-tailed t-tests for each inhibitor-exposed sample with respect to the control. p-values from left to right: 0.25, 0.0398, 0.0199, 7.9 × 10−4.