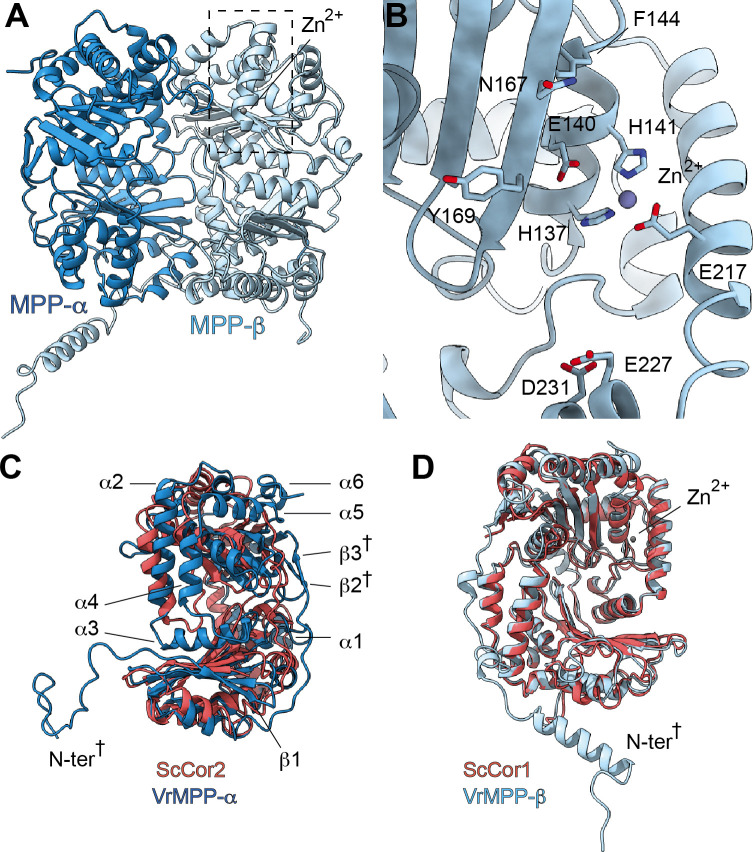
Figure 3. V. radiata’s CIII2 mitochondrial processing peptidase (MPP) domain has a conserved architecture and active site but contains plant-specific secondary-structure elements not seen in other CIII-MPP subunits or in soluble MPP.
(A) Ribbon representation of the VrMPP-α (blue) and VrMPP-β (light blue) looking into the central cavity. Dashed rectangle indicates the location of the active site, detailed in (B). (B) MPP-β active site [rotated 90° about vertical axis with respect to (A)]. Shown in stick representation are the Zn-coordinating residues (His137, His141, Glu217), the catalytic water-activating residue (Glu140) and conserved, putative substrate-recognition residues (Phe144, Glu227, Asp231, Asn167, Tyr169). Residue Ala168 is also conserved, but not visible in this orientation. (C–D) Superposition of V. radiata and S. cerevisiae CIII2 MPP domain subunits. VrMPP-α and -β’s structural elements not present in ScCor2 and ScCor1 are marked. Structural elements that are additionally not present in yeast soluble MPP, i.e. plant-specific features, are marked with a cross (†). (D) VrMPP-α (blue) and ScCor2 (dark pink). (D) VrMPP-β (light blue) and ScCor1 (dark pink). β, β−strand; α, α-helix; N-ter, N-terminus. See also Figure 3—figure supplements 1–2 for further details. S. cerevisiae structures from PDB: 6HU9.