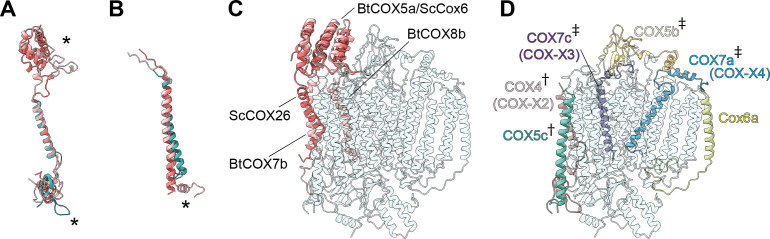
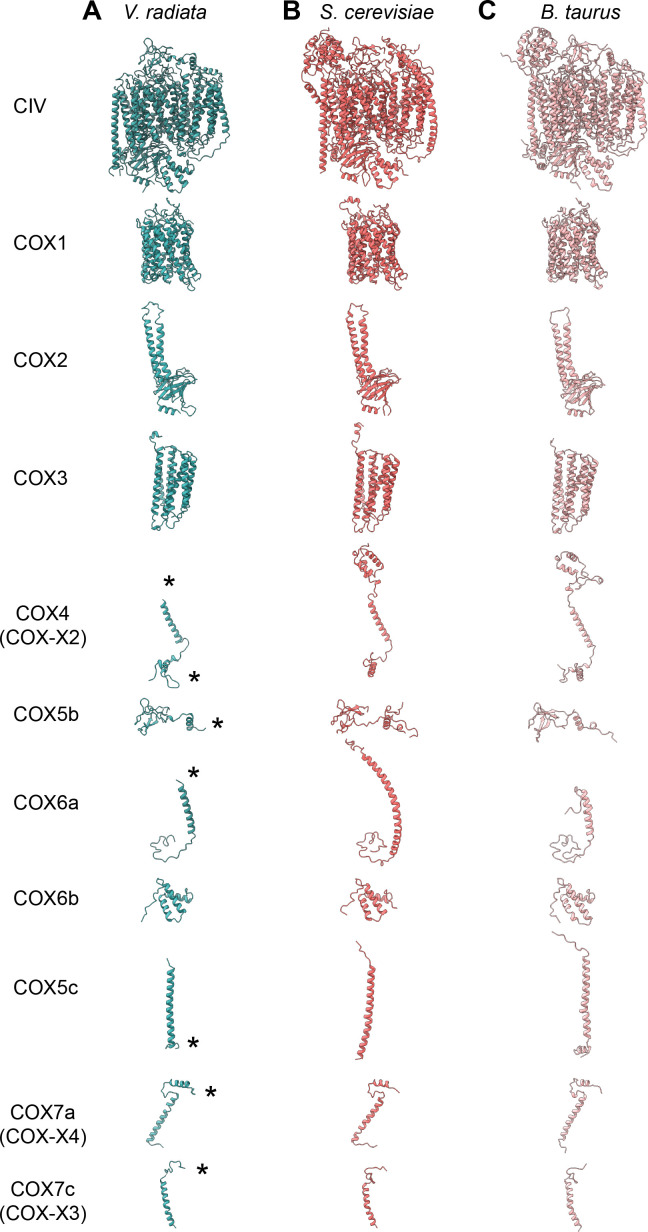
Figure 6. Subunit differences in V. radiata CIV.
(A–C) Superposition of subunits of V. radiata (Vr, teal), S. cerevisiae (Sc, dark pink; PDB: 6HU9) and B. taurus (Bt, light pink; PDB: 5B1A) CIV. Subunits were aligned with the corresponding V. radiata subunit. Asterisks mark the differences discussed in the text. Names of the subunits previously believed to be plant-specific are given in parentheses. (A) Superposition of VrCOX4 (COX-X2), ScCox5a, BtCOX4. (B) Superposition of VrCOX5c, ScCox9, BtCOX6c. (C) Superposition of the yeast and bovine CIV subunits that do not have homologs in V. radiata, onto the V. radiata CIV model (transparent teal). Alignment by COX1 subunits. (D) Location of the V. radiata accessory subunits that show notable differences with their yeast/bovine homologs. Subunits that form the supercomplex interface in V. radiata are marked with (†). Subunits whose homologs form the supercomplex interface in the B. taurus respirasome are marked with (‡).