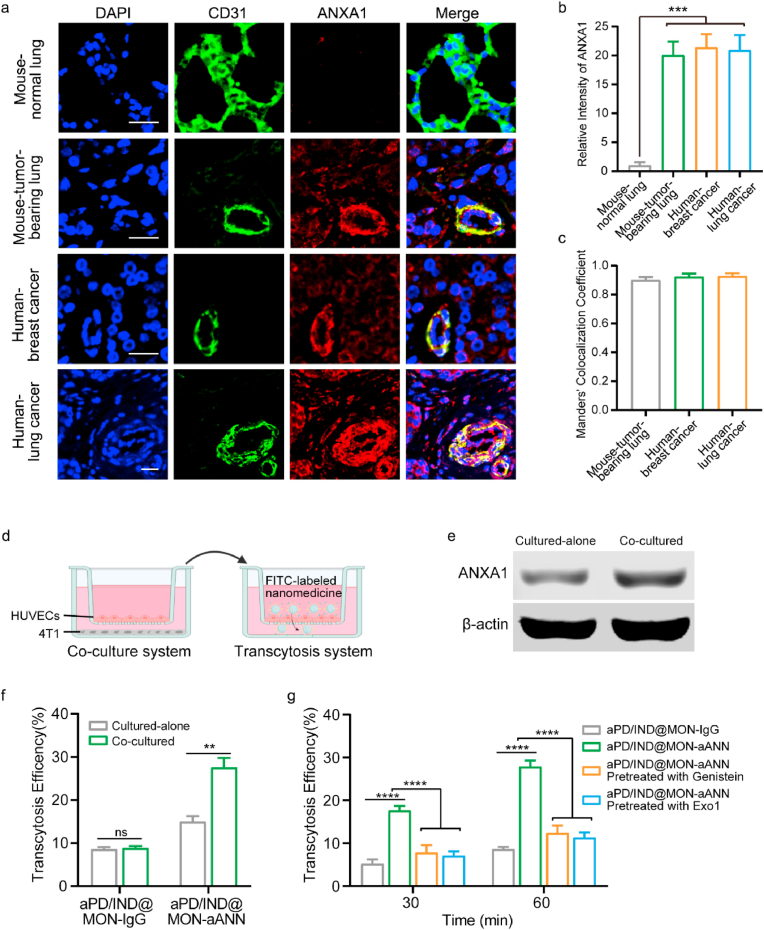
Fig. 2.
in vitro nanoparticle transcytosis across tumor endothelial cells. (a) Representative immunofluorescence images of CD31 (green) and ANXA1 (red) of various tumor tissue samples with the colocalization shown in yellow color. Scale bar = 20 μm. (b) Quantitative analysis of ANXA1 fluorescence intensity in the CD31 positive zone for various tissue samples (n = 9 images from three independent experiments, data presented as mean ± s.d, one-way ANOVA with Tukey test, ***P < 0.001). (c) Quantitative analysis of co-localization between CD31 and ANXA1 fluorescent signals in the tumor tissue sections, Manders' Colocalization Coefficient denotes the fraction of CD31 overlapping with ANXA1. The coefficient is close to 1 when they are highly co-localized (n = 9 images from three independent experiments). (d) Illustration of the co-culture system between HUVECs and 4T1 tumor cells and transcytosis assay model. (e) Representative western blotting result of ANXA1 in HUVECs cultured alone or co-cultured with 4T1 cells. (f) Transcytosis efficiency of FITC-labelled aPD/IND@MON-IgG and aPD/IND@MON-aANN at 60 min in the transcytosis model with HUVECs cultured alone or pretreated with 4T1 co-culture (n = 3, data presented as mean ± s.d, one-way ANOVA with Tukey test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (g) Transcytosis efficiency of FITC-labelled aPD/IND@MON-IgG and aPD/IND@MON-aANN at 30 min and 60 min in the transcytosis model pretreated with or without 200 μM genistein (endocytic pathway inhibitor) or 20 μM EXO1 (exocytic pathway inhibitor) (n = 3, data presented as mean ± s.d, one-way ANOVA with Tukey test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).