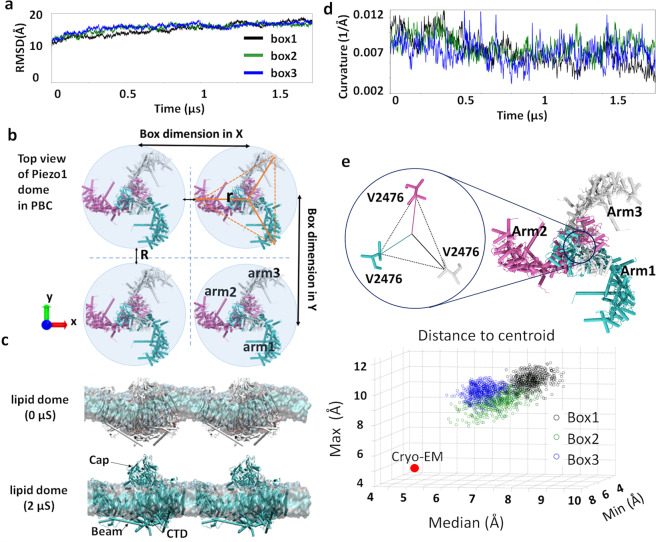
Fig. 2. Three 1.75 µs Piezo1 all-atom MD simulations.
a Protein backbone RMSD in three systems, named box1, box2, and box3. b Piezo PBC images along x-axis and y-axis. Arm 1 in cyan, arm2 in mauve, and arm3 in white. The centroid distance used to calculate the radius of the Piezo1 dome (r) is illustrated using solid orange lines. The dome–dome distance (R) and the box size in x- and y-dimensions are shown using black double arrow lines. c Flattening of the lipid dome between two Piezo1 channels, illustrated by the snapshots at 0 and 2 µs of AA simulation of the box1 system. d Mean curvature of three systems from fitting lipid headgroups to a 2D Gaussian model (see “Methods” section for details and code). e Scatter plot of three V2476 centroid distances (min, median, and max). The circle symbol for each system marks the data points from the last 500 frames (60 ns). The red dot marks the corresponding valine centroid distance from the cryo-EM structure. The triangle formed by carbon β of V2476 residues from three subunits is shown on the top.