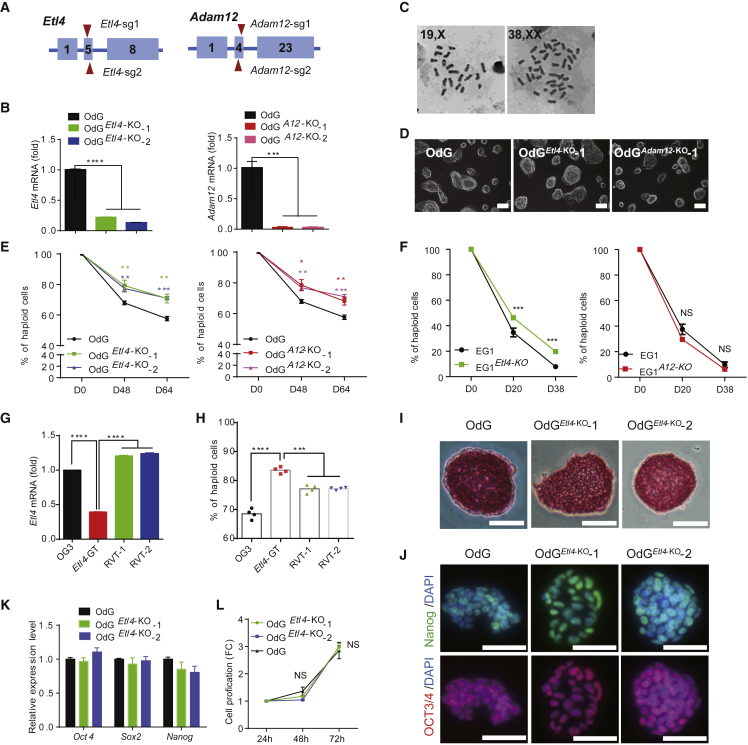
Figure 2.
Etl4 Deficiency Facilitates the Maintenance of Mouse haESCs
(A) Schematic diagram of the strategy to knockout Etl4 and Adam12 in OdG haESCs by using CRISPR/Cas9 system. Boxes indicate the exon of the genes and the triangles represent the single guide RNA (sgRNA). The sequences of the sgRNAs are listed in Table S1.
(B) Validation of Etl4 and Adam12 expression level in mutant and WT OdG haESCs by RT-qPCR. The expression level was normalized by Gapdh (n = 3 independent experiments).
(C) Chromosome spreads of Etl4-KO OdG haESCs showing normal haploidy. The right panel depicts the self-diploidized Etl4-KO cells.
(D) Morphology of Etl4 and Adam12-deficient OdG haESCs cultured in M2iL medium. Scale bars, 100 μm.
(E and F) Flow cytometry analysis of the ratio of haploid cells in (E) Etl4-KO, Adam12-KO and (F) two corresponding WT haESC lines during cell passages.
(G) Expressional level of Etl4 in OG3, Etl4-GT, RVT-1, and RVT-2 haESCs tested by RT-qPCR (n = 3 independent experiments).
(H) Percentages of haploid cells in OG3, Etl4-GT, RVT-1, and RVT-2 haESCs after 12 days of culture (n = 3 independent experiments).
(I) Alkaline phosphatase activity in Etl4-KO and WT OdG haESCs. Scale bars, 50 μm.
(J) Immunofluorescence (IF) staining for the indicated pluripotency markers in Etl4-KO and WT OdG haESCs. Scale bars, 50 μm.
(K) Validation of indicated pluripotency-related markers' expression level in Etl4-KO and WT OdG haESCs by RT-qPCR. The expression level was normalized by Gapdh (n = 3 independent experiments).
(L) Cell proliferation rate of Etl4-KO and WT OdG haESCs (n = 3 independent experiments). Data in (B), (E), (F), (G), and (H) are shown as mean ± SEM, ∗p < 0.05 and ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. NS, not significantly different.