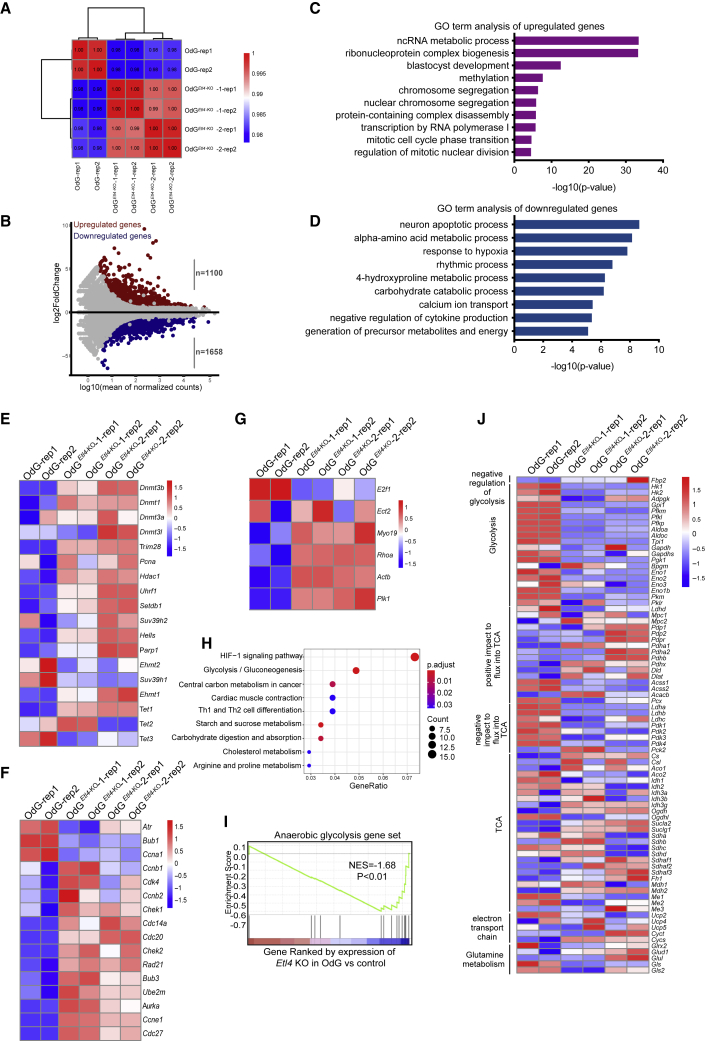
Figure 3.
Transcriptome of Etl4-deficient OdG haESCs
(A) Hierarchical clustering of gene expression profiles from two biologically independent samples based on Pearson correlation coefficient in two Etl4-KO and WT haES clones. Colors from green to red indicate weak to strong correlations.
(B) MA plots showing gene expression changes in Etl4-KO haESCs. Red dots indicate upregulated genes and blue dots indicate downregulated genes.
(C and D) GO analysis of upregulated genes (C) and downregulated genes (D) for biological processes in Etl4-KO OdG haESCs.
(E–G) Heatmap showing the expression of methylation related genes (E), cell-cycle–related genes (F), and Rho A pathway (G) in Etl4-KO and WT OdG haESCs.
(H) Significantly enriched KEGG pathways for genes expressed differentially in Etl4-KO OdG haESCs.
(I) GSEA for anaerobic glycolysis gene set in WT OdG and Etl4-KO haESCs. For the x axis, genes were ranked based on the ratio of Etl4-KO versus WT haESCs.
(J) Heatmap showing changes in RNA expression levels for various enzymes and regulators of central carbon metabolism in Etl4-KO and WT OdG haESCs. The scale represents Z score.