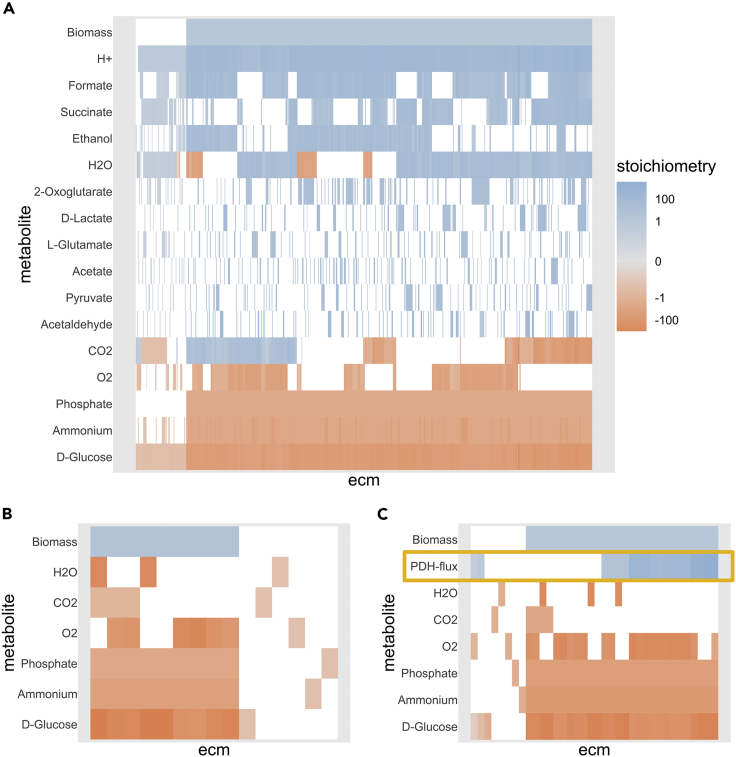
Figure 3.
The Full Metabolic Potential of the e_coli_core-Model
(A) The ECMs of the full model are shown as the different columns; each row corresponds to a different external metabolite. The color scale indicates the stoichiometric coefficient of the metabolite in the conversion: blue for production and red for consumption. The coefficients were log-transformed to allow for visualization of differences in both large and small coefficients (details and R-code can be found in Supplemental Information 10.2); small values are shown in gray while zero values are white. Of the 689 elementary conversions, 613 lead to the production of biomass. These ECMs were normalized to fix the biomass production at 1, while the other ECMs were normalized such that the sum of absolute coefficients is 1.
(B) If we use the hide method, explained in Box 2, to hide the production of metabolites, we get 15 ECMs that span all possible ratios in which substrates can be converted into biomass. This smaller set of ECMs is easier to compute and easier to explore, while the steady-state assumption is still satisfied in the whole network. So even though the secretion of products is not reported, it has been implicitly taken into account, so that all relations between substrates shown in (A) are captured in (B).
(C) If we use the tag method, also explained in Box 2, to report the activity of the pyruvate dehydrogenase (PDH) reaction, we find 36 ECMs that summarize all possibilities. It can be seen that the PDH reaction is not essential for growth but seems to be necessary for efficient growth on glucose, since the uptake of glucose is generally lower when PDH is active.