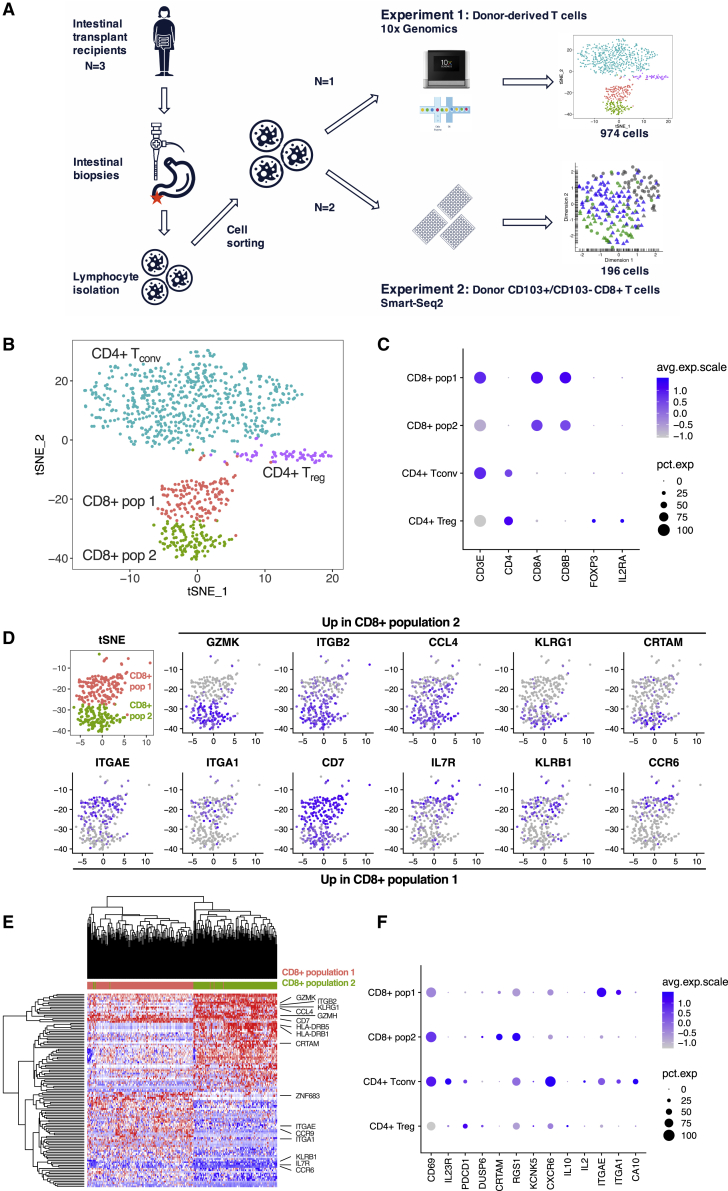
Figure 2.
Single-cell RNA sequencing (scRNA-seq) delineates transcriptionally distinct states within CD4+ and CD8+ TRM cell populations
(A) Schematic of two scRNA-seq experiments. Biopsies of small intestinal transplant tissue were collected from subjects at endoscopy, then dissociated to isolate intestinal lymphocytes. These were sorted by fluorescence-activated cell sorting (FACS), first with bulk sorting of donor-derived T cells in experiment 1, then with index sorting of donor-derived CD103− and CD103+ T cells in experiment 2, before scRNA-seq library preparation and sequencing using the 10× Genomics platform (experiment 1) or the Smart-Seq2 protocol (experiment 2).
(B) t-Distributed Stochastic Neighbor Embedding (tSNE) plot of 974 donor-derived T cells from a single subject showing four transcriptionally distinct populations of conventional CD4+ and CD8+ T cells.
(C) Dot plot of key gene identifiers for the four clusters showing two clusters of CD8+ T cells and two clusters of CD4+ T cells, including one containing cells expressing IL2RA and FOXP3, consistent with a regulatory T cell phenotype. Dot size indicates the proportion of cells in which the gene is expressed.
(D) tSNE plots showing the expression of key genes upregulated in CD8+ population 2 (top row) or upregulated in CD8+ population 1 (bottom row).
(E) Heatmap indicating hierarchical clustering of gene expression of CD8+ donor-derived T cells from population 1 and population 2. Cell labels above indicate cluster: red, population 1; green, population 2. Genes of interest are highlighted.
(F) Dot plot showing the expression of 13 genes previously associated with tissue residency in human CD69+ T cells (Kumar et al., 2017). Dot size indicates the proportion of cells in which the gene is expressed.