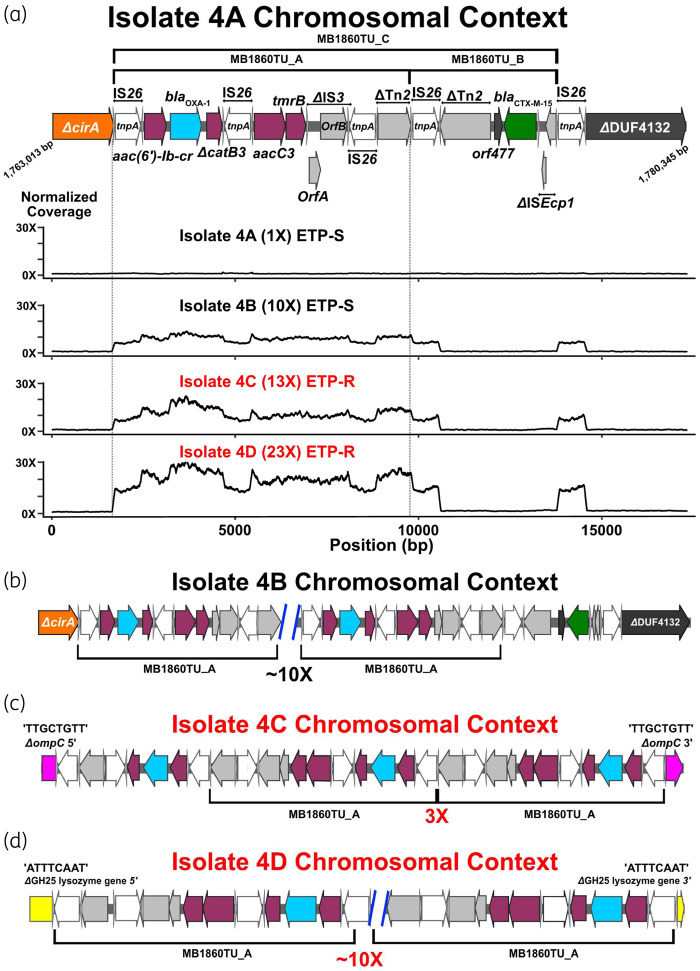
Figure 3.
Characterization of IS26-flanked transposon, TnMB1860, with transposition of modular TUs in Patient 4 serial isolates (i.e. isolates 4A–D). Terminal left and right inverted repeats (IRL and IRR, respectively) of ISs are specified by grey triangles that bracket complete and incomplete tnpA genes, respectively. ORFs are coloured as follows: non-β-lactamase AMR genes (maroon), blaOXA-1 (blue), blaCTX-M-15 (green), IS26 tnpA (white) and other IS/Tn elements (grey). Font colour for each serial isolate labelled represents ertapenem susceptibility (black) or ertapenem resistance (red). (a) Schematic indicates chromosomal context (GenBank accession #: CP049085) of TnMB1860 locus flanked by directly oriented IS26 transposases found in the 4A isolate. Immediately below the schematic are normalized, short-read coverage depth line graphs for the four Patient 4 serial isolates with MB1860TU_A bracketed by dotted lines. ETP-S, ertapenem susceptible; ETP-R, ertapenem resistant. (b–d) Characterization of MB1860TU_A transposition and ORF disruption events in each of the respective Patient 4 recurrent episode isolates. Black brackets beneath ORFs indicate MB168TU_A. (b) Isolate 4B chromosomal context shows an ∼10× MB1860TU_A amplification event in the original 4A chromosomal locus. (c) Isolate 4C chromosomal context additionally contains transposition and disruption of the ompC porin gene (pink) upstream of the original isolate 4A TnMB1860 locus with three copies of MB1860TU_A. (d) Isolate 4D has previous transposition events found in isolates 4B and 4C as well as another MB1860TU_A transposition, amplification and disruption of a putative glycoside hydrolase gene (i.e. GH25; yellow) downstream of the TnMB1860 locus. The TSDs created by the transposition of the TU are indicated above each respective junction site flanking the insertion and disruption of the ompC and GH25 genes, respectively.