Figure 2. SG condensation requires G3BP-UBAP2L complexes.
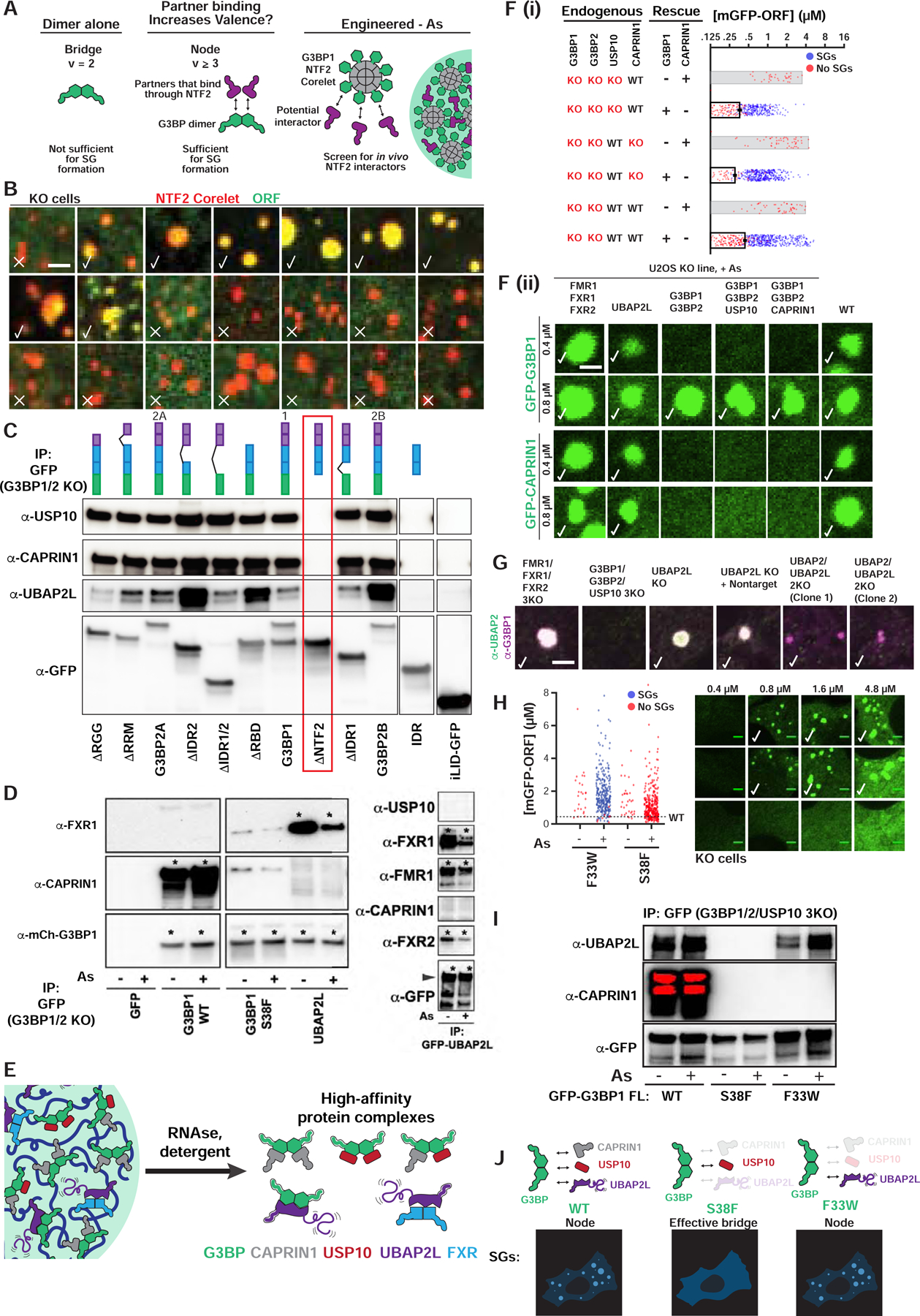
(A) Dimeric G3BP RBD bridges (v=2) are not sufficient for SGs; G3BP must act as node (v>2) via additional high-affinity protein-protein interactions (PPIs) with its NTF2 dimerization domain; right: live cell Corelet assay to screen for PPIs.
(B) G3BP KO cells (No As) with G3BP NTF2 Corelets (red, sspB-mCh-G3BP1ΔRBD; no tag, iLID-Fe core) and GFP-tagged proteins (10-min activation). Checks = putative NTF2 partners/PPIs.
(C) GFP-G3BP1Δs IPed from G3BP KO cells (No As) with α-GFP (then RNase and RIPA-wash) to isolate tightly-bound proteins. ΔNTF2 (red box) abolishes binding. Representative blot (n=3 experiments).
(D) GFP-tagged proteins IPed similar to (C), but +/− As. Representative blot (n=3 experiments), * = high-affinity interaction.
(E) High-affinity, RNA-independent complexes predicted by IPs.
(F) Top (i): Quantification of GFP-G3BP concentration threshold for SGs in KO cells (+As). Mean and SEM: n=3 experiments (n>4 images per). Bottom (ii): KO cells with GFP-tagged protein at indicated concentration, check = SGs, check* = smaller SGs.
(G) Panel of U2OS KO cells (+As) examined for SGs by immunofluorescence. Indicated: no SG defect (check), smaller SGs (check*), very small SGs in rare cells (check**).
(H) Quantification of G3BP variant concentration threshold for SGs in G3BP KO cells (+/− As). Mean and SEM: n=3 experiments (>4 images per). Representative images at indicated concentrations (+As, check=SGs).
(I) GFP-G3BP variants IPed similar to (D), but in G3BP1/2/USP10 3KO cells. Representative blot (n=3 experiments).
(J) G3BP variants form complexes of different valence, which corresponds to ability to rescue SG defects.
See also Figure S2.
