Figure 3. Valence capping of the G3BP node by RBD-lacking binding partners prevents stress granule formation.
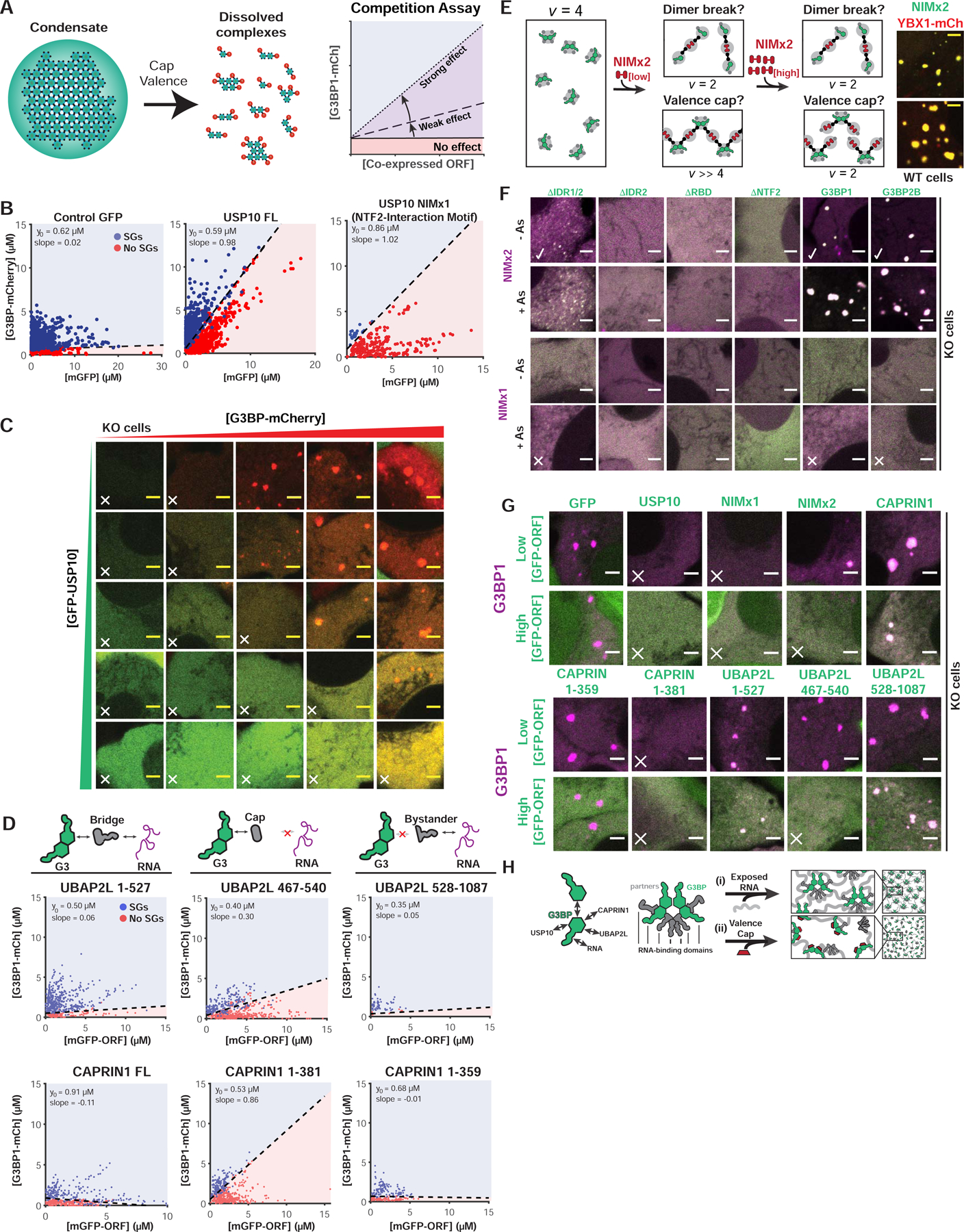
(A) Interacting “caps” (v=1) are proposed to disrupt networks of high v particles. Right: SG rescue competition assay (G3BP KO cells) tests model by co-expressing GFP-tagged NTF2 partners (cap, positive slope) with G3BP1-mCh.
(B) Competition assay for predicted caps in G3BP KO cells (+As). Indicated: y-intercept (G3BP rescue concentration, no competitor), best-fit slope demarcating +/− SG cells.
(C) Representative images for (B, middle) at indicated protein concentrations (X, no SGs).
(D) Competition assay similar to (B) with CAPRIN1/UBAP2LΔs.
(E) NTF2-interacting motifs (NIMs) inhibit SGs by “dimer breaking” or “valence capping”, differentiable using a v=2 NIM bridge (“NIMx2”). If capping: low NIMx2 promotes condensation, polymerizing G3BP dimers (high vRBD); high, inhibits by saturation (vRBD=2). If breaking, low and high NIMx2 link G3BP monomers (vRBD=2). Right: GFP-NIMx2 induces SGs in WT U2OS (-As).
(F) Representative images (X, inhibits SGs; check, promotes): G3BP KO cells (+/− As) expressing GFP-G3BPΔs and mCh-NIMx1 (or x2)
(G) Images (X, inhibits SGs) for G3BP KO cells (+As) with mCh-G3BP1 and GFP-tagged protein (low or high levels).
(H) Molecular model for SG regulation by NTF2 PPIs.
See also Figure S3.
