Figure 5. Stress granules with attached P-bodies are the default multiphase condensate encoded by high valence RBD nodes.
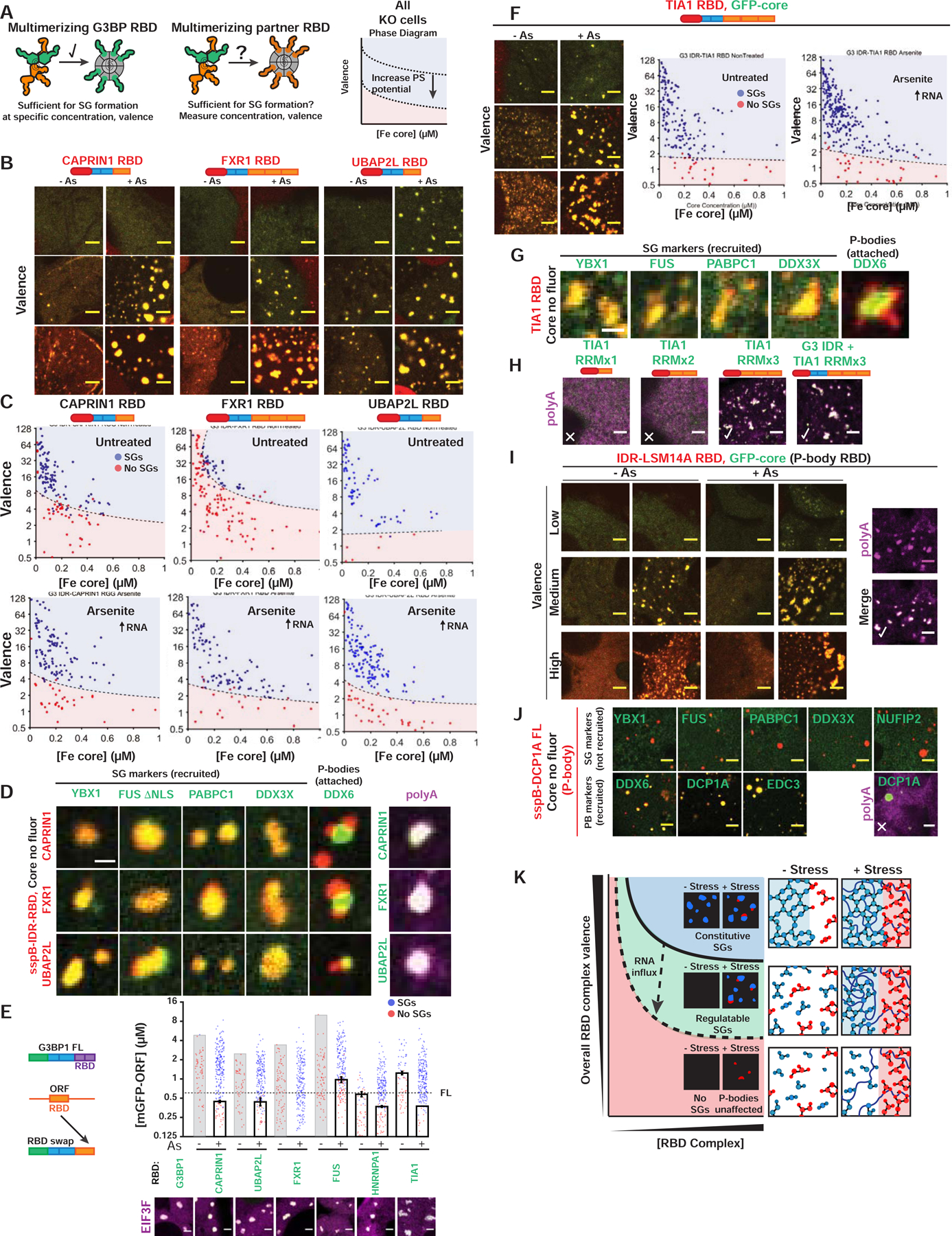
(A) Corelet assay to test whether NTF2 partners contribute vRBD to G3BP complex.
(B) Valence-dependent condensation (+/−As) examined for indicated RBDs fused to G3BP IDR in Corelet system (images correspond to (C)). All Corelet experiment images (unless noted): vRBD is noted low (~2–4), medium (~6–8), or high (~18–24); core ~0.25 μM; cells = G3BP KO U2OS; scale bar = 3 μm.
(C) Intracellular phase diagrams for RBDs in (B) +/− As. Each dot = single cell (5-min activation), best-fit phase threshold shown.
(D) GFP-tagged proteins expressed with indicated RBD Corelets (iLID-Fe lacks GFP tag). Following As and 10-min activation, cells were fixed; arrowheads, PBs attached to SGs. Right: oligo-dT RNA FISH.
(E) SG rescue threshold for GFP-tagged chimeric G3BP1 with swapped RBDs (G3BP KO cells with EIF3F-mCh, representative images below). Mean and SEM: n=4 experiments (>4 images per).
(F) Similar to (B,C) but with TIA1 RBD Corelets.
(G) Similar to (D) but with TIA1 RBD Corelets.
(H) Similar to (D) but with TIA1 RBD (number of RRMs altered; +/− G3BP1 IDR).
(I) Similar to (B,D) but with RBD from LSM14A (essential PB protein).
(J) Similar to (D) but with DCP1A (PB protein that lacks RBD).
(K) Phase diagram cartoon depicting SG formation as function of nucleating complex concentration and its vRBD. WT cells would exist in green region; G3BP KO/capped, red.
See also Figure S5.
