Figure 3. Temporal Resolution of SG Disassembly Reveals a Network of DEPs.
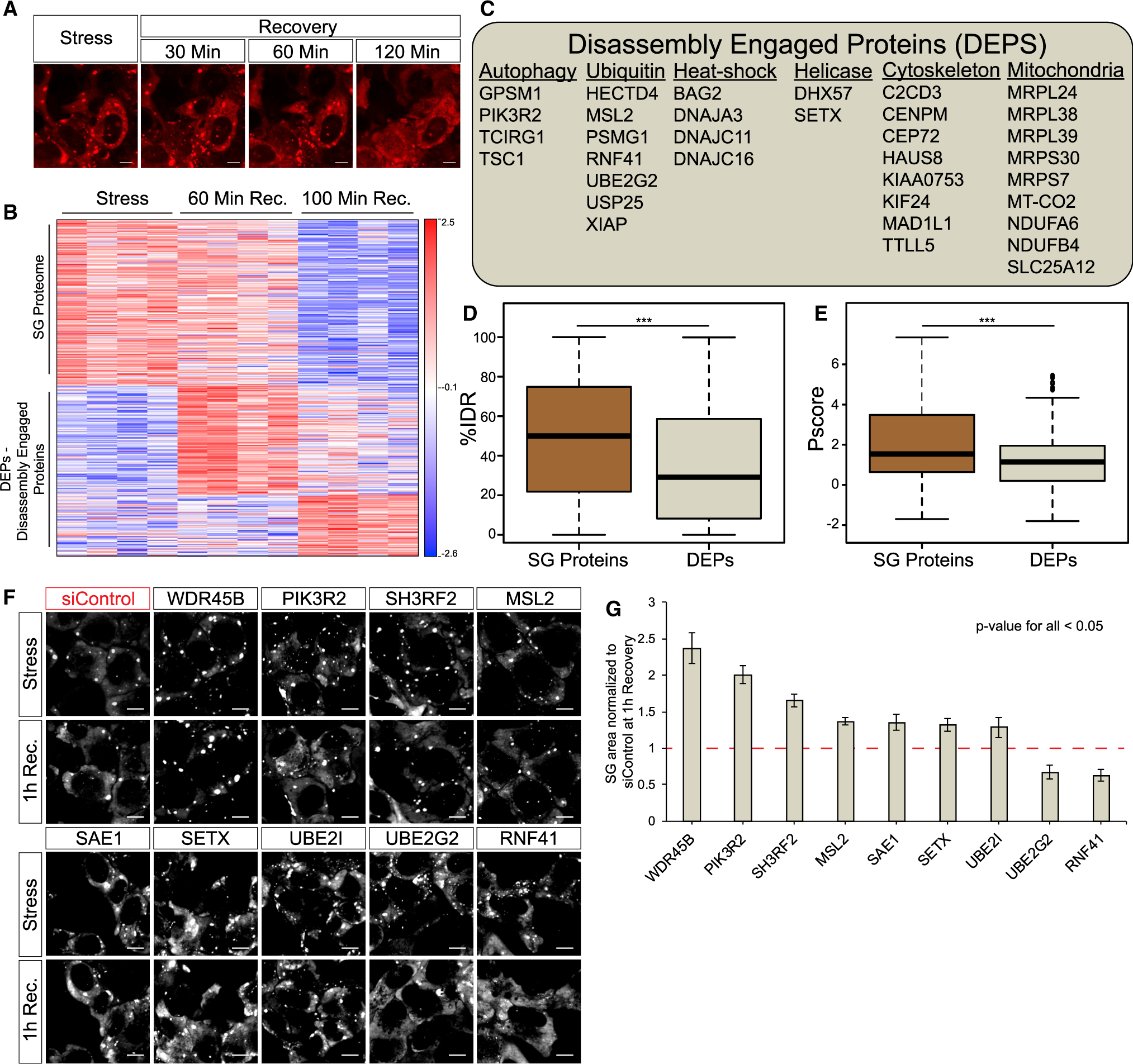
(A) Representative micrographs depicting RFP-G3BP1 in stressed U2OS cells and 30, 60, and 120 min during recovery, after the stressor was washed out; scale bar, 10 μm.
(B) Heatmap of unsupervised clustering of proteins associated with SG disassembly. Proteins specifically enriched in SG relative to the cytoplasm, if exceeding a 2-fold enrichment in FMR1-APEX/NES-APEX and p < 0.05 by Student’s t test with correction to multiple hypothesis by FDR. 224 proteins are enriched in SGs, while 202 proteins are enriched in SGs once stress is removed and disassembly dynamics ensue.
(C) A list of representative disassembly-engaged proteins (DEPs) associated with different cellular pathways.
(D) Boxplot analysis of IDR enrichment (%IDR; Mészáros et al., 2018) in 224 SG resident proteins or 202 DEPs. Upper and lower quartiles and extreme points are shown. Wilcoxon signed-rank test p < 0.0005.
(E) Propensity to phase separate (Vernon et al., 2018) in 224 SG resident proteins or 202 DEPs. Upper and lower quartiles and extreme points are shown. Two-sided Student’s t test p < 0.0005.
(F) Representative micrographs depicting GFP-G3BP1 in stressed U2OS cells and during recovery after the stressor was washed out. Cells were transfected with non-targeting siControl or specific siRNAs for the knockdown of DEPs; scale bar, 10 μm.
(G) Graph quantification of SG disassembly dynamics by live GFP-G3BP1 imaging. SG area, normalized to siControl area (y axis) at 1 h after stress washout. Three experimental repeats for measurement with four different areas per well. Representative experiment from two independent live-imaging studies tested by ANOVA repeated-measurement, p < 0.05. siRNA tested that failed statistical significance were Topors, Becn1, Bag2, Kif24, Mex3c, Ranbp2, Nbr1, Hsbp1, Dst, Cep72, Xiap, Usp25, and Traf5.
