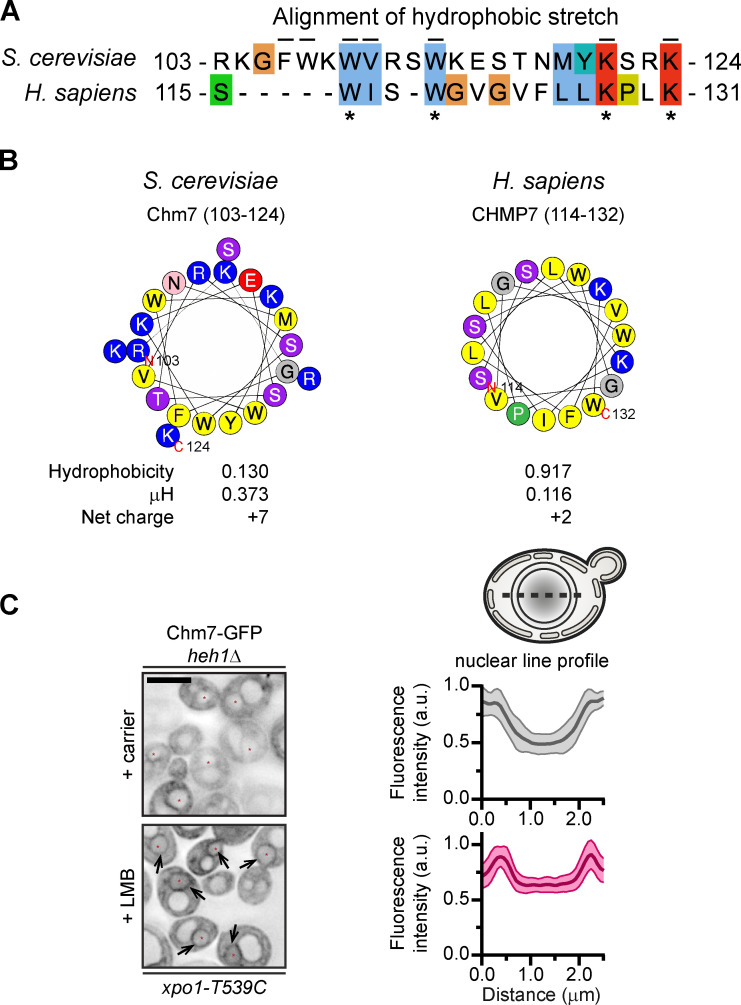
Figure S1.
Conserved hydrophobic stretch within Chm7 can be modeled as an amphipathic helix, and Chm7 accumulates at the INM without Heh1 (supplemental to Fig. 1). (A) Alignment of hydrophobic stretches in budding yeast and human. Color coding of properties of amino acids with conserved attributes as in ClustalW (Larkin et al., 2007), where hydrophobic residues are blue, positive-charged residues are red, polar residues are green, glycines are orange, prolines are yellow, and aromatic residues are cyan. Numbers are amino acids. Asterisks denote conserved residues; black lines are residues altered in mutational analysis in this study for Chm7 (top). (B) Helical wheel diagrams in which the letters are amino acids. Physicochemical properties were calculated by using HeliQuest (Gautier et al., 2008). Residues are color coded where hydrophobic residues are yellow, basic residues are blue, serine and threonine are purple, acidic residues are red, asparagine is pink, glycine is gray, and proline is green. (C) Chm7 accumulates at the INM in the absence of Heh1. Deconvolved inverted fluorescence micrographs of overexpressed Chm7-GFP in an heh1Δ LMB-sensitive strain (xpo1-T539C) after treatment with carrier (MeOH) or LMB. Arrows point out Chm7-GFP that accumulates at the nuclear rim specifically under conditions of LMB treatment. Scale bar, 5 µm. At right are line profiles bisecting the nucleus of carrier and LMB-treated cells where the mean Chm7-GFP fluorescence intensity (in a.u.) is plotted. The mean (normalized to max) is shown as thick lines and SD as thin lines from 20 randomly selected cells. Gray is the + carrier (control) treatment, and magenta is the +LMB treatment.