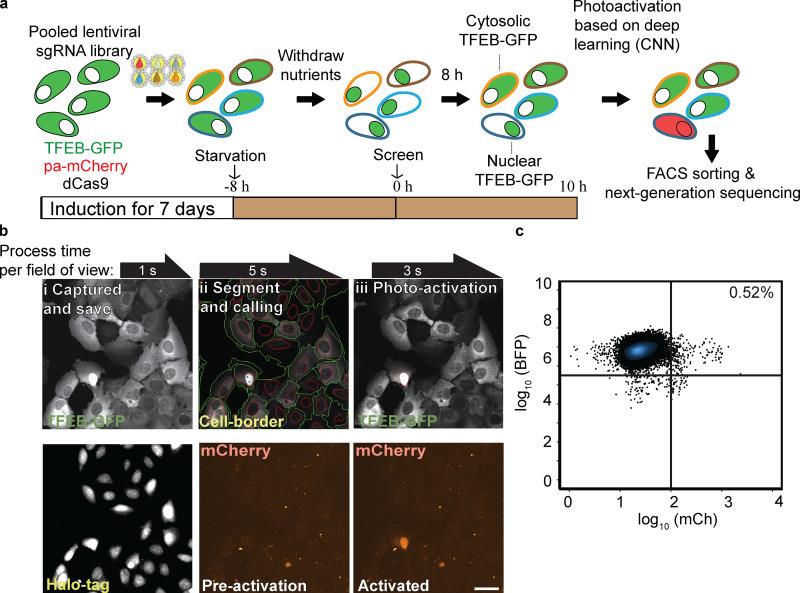
Figure 5.
Whole-genome GFP-TFEB localization screen. (a) Screen workflow. GFP-TFEB cells were transduced with pooled sgRNA libraries targeting the whole genome for 7 d. Following 8 h of starvation, AI-PS screening platform was initiated. (b) Images examples of field of view of GFP-TFEB U2OS cell-screening procedure. i: Images were captured and saved on a local computer. Top: GFP-TFEB; bottom: 2 µM of JD ligand Halo-tag ligand were admitted for nucleus staining. ii: Cells borders were identified (green circle surrounding cell border, red circle the nucleus) following nucleus segmentation. Bottom: mCherry channel for ph-mCh protein. iii: CNN classification model was deployed and masked (red circle). Photoactivation of the CNN identified cell. Scale bar, 10 µm. (c) Flow cytometry sorting of 249 cells from 46,981 (flow cytometry raw data and gating instruction is stored on GitHub). Flow cytometry scatterplot representing the separation of the postscreen photoactivated from the inactivated cell population. BFP florescence signal y-axis in cyan, mCherry florescence signal x-axis in red. For three biologic repeats of seven subpooled libraries (composing the whole genome), ∼12,600,000 cells were screened and 25,579 cells were photoactivated sorted and sequenced.