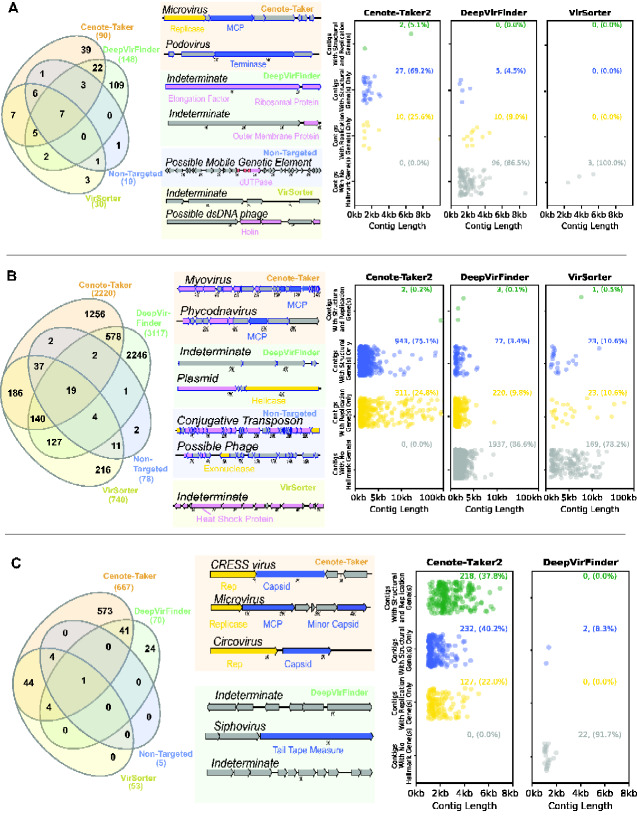
Figure 3.
Comparison of virus discovery tools for DNA datasets. Contigs >1,000 nucleotides were analyzed using four virus detection/discovery pipelines. (A) A dataset for human stool enriched for nuclease-resistant DNA in virus-sized particles (SRR6128021). Left panel: Venn diagram displaying the overlap of contigs identified as viruses by the various pipelines. Middle panel: maps showing representative examples of unique calls from each pipeline. Left panel: scatter plots display the length distribution for calls unique to each software package. Rows of plots show contigs that contain both virion structural genes and DNA or RNA replication-associated genes, only one of the two categories of gene, or neither. (B) Similar display of a dataset for Amazon River water samples analyzed with shotgun total DNA sequencing (ERR2338392). (C) Similar display of a dataset for wastewater enriched for nuclease-resistant DNA in virus-sized particles amplified using MDA/RCA (SRR3580070).