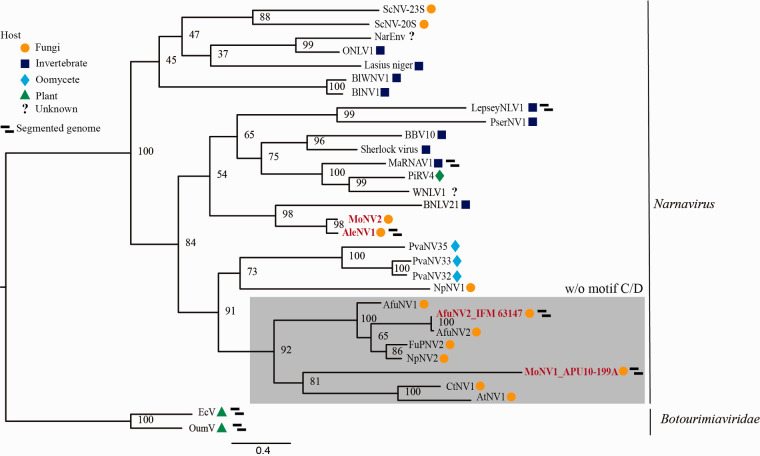
Figure 3.
Phylogenetic tree for the RdRp from the Narnaviridae family. The RdRp amino acid sequences without motifs C and D from AleNV1, AfuNV2, MoNV1, and MoNV2 (without the C/D motif is shown in a gray box) and related viruses were used to construct the phylogenetic tree. The numbers indicate the percentage bootstrap support from 1,000 RAxML bootstrap replicates. The best-fitting amino acid substitution model was [LG + F + G]. The accession numbers and full virus names are listed in Table S4. The viral sequences from fungi, invertebrates, oomycetes, plants, and an unknown host are indicated by orange circles, blue squares, light blue diamonds, green triangles, and question marks, respectively. The viral sequences identified in this study are shown in red font.