Abstract
Background:
Campylobacter spp. are the main cause of human gastroenteritis. The 16SrRNA sequencing is one of fast molecualr method to detect this fastidious. In this study, we compared the sequencing of 16srRNA genewith four housekeeping genestodetect Campylobacter spp. in patients with diarrhea and healthy people.
Methods:
60 samples of Campylobacter DNA extracted from stool samples of 30 patients with diarrhea and 30 healthy people were used. In order to detect Campylobacter, we designed primers for proliferation of 16SrRNA, cadF, dnaJ, slyD , and rpoA genes using Primer 3, Mega 4.0 and Blast software. Then the PCR products were sequenced using ABI system.
Results:
The sequencing showed concordance of PCR-products with deposited sequences in the Gene Bank. Among diarrhea patients, 53.3% of samples were significantly (p< 0.05) positive for slyD and cadF genes and 50% of samples were positive using 16SrRNA, rpoA, and dnaJ genes by PCR assay. The average of sensitivity and specificity were found 53.33% and 83.33%, respectively.
Conclusion:
Due to various copies of repeated sequences of 16SrRNA gene, analyzing its amplicons on electrophoresis may be more difficult than the slyD and cadF genes. According to our results, among the 5 studied genes; the highest detection rate was related to slyD and cadF genes. Although, dnaJ and rpoA genes,instead of 16SrRNA gene, can be considered as appropriate genes for molecular detection of Campylobacter bacteria.
Keywords: cadF, Campylobacter, Diarrhea, Molecular detection, slyD
Introduction
Campylobacter spp. are gram-negative, spiral,and coma-shape bacteria. Campylobacter spp. require microaerophilic conditions (10% CO2,5% O2) and also temperatures 37- 42 °C to grow (1). The size of Campylobacter genome is 1.6 to 7.1 MBp (1). Virulence factors of Campylobacter such as motility, adhesion, invasion to host cell, and production toxin contribute in pathogenicity and causing host cell necrosis (3,4). Among virulence genes of Campylobacter, flaA, flaB,racR, dnaJ, and, slyD are known to be responsible for pathogenesis, while ciaB and pldA contribute in gene expression and colonization. Also, cdtA, cdtB, and cdtC genes are known to be responsible for production of cytotoxin (5). The slyD gene encodes Peptidyl Prolyl cis/ trans-isomerase which has role in the synthesis of amino acids(6). Small subunit ribosomal RNA (16SrRNA) gene is one of the most important target genes in molecular studies on the bacterial evolution and epidemiology (7). The 16SrRNA gene is considered as a gold standard gene for estimating of phylogenetic diversity in microbial communities (8,9); Nevertheless, the widespread usage of 16SrRNA, it can limit several aspects of the results as its high copy numbers in the genome, 1-15 or more copies (10). The copy numbers can Downloaded from differs between Campylobacter spp. and only a limited number of bacterial genomes have one copy of 16SrRNA gene that its varies may be simultaneous with increasing number of copies (11). Also, the sequence of 16SrRNA gene can be different among bacteria communities (12).
One of the main causes of diarrhea in humans is Campylobacter infection and its detection using bacterial culture is not easy. Therefore, accurate identification of this gene by molecular methods is very important.
The purpose of this study is to compare the detection of Campylobacter using the 16SrRNA gene with slyD, cadF, rpoA, and dnaJ genes for determination in patients with diarrhea admitted to Amirkabir Hospital in Arak and healthy individuals people referred to the Arak health center in 2017-2018.
Materials and Methods
Primary isolation
This study was approved by the ethics committee of Arak Universityof Medical Sciences (Ethics code:IR.ARAKMU.REC.1397.229). We used 60 DNA samples available in DNA Bank of Infectious Diseases Research Center of Arak University of Medical Sciences, Iran. DNA samples were extracted from human stools and included 30 samples of patients with diarrhea and 30 fecal specimens from healthy individuals exposed to poultry meat. Because the chicken is a major host for the transmission of Campylobacter species.
Detection of Campylobactergenus by Polymerase chain reaction (PCR) and Sequencing
For each sample, five housekeeping genes, 16SrRNA, cadF, rpoA, dnaJ , and, slyD were used in PCR. Properties of primers are shown in Table 1. The reaction mixture in final volume was 15 μl, contained 6.2 μl of Master mix (ID No: 5200300-1250, YTA, Iran), 50 ng genomics DNA, and 0.7 μl (10 pmol) of each primer (Copenhagen, Denmark). The PCR was performed in Thermocycler device (Eppendorf, Germany) under the following temperature conditions: denaturation at 95 °C for 5 min, 35 cycles including initial denaturation of 95 °C for 1 min, annealing 56.4°C (16SrRNA gene) for 55 sec, Extension 72 °C for 1 min and final Extension 72 °C for 10 min. Annealing temperature for each gene is provided in Table 1. DNA extracted from Campylobacter colonies (from previous study Reference?) and distilled water were used as positive and negative control, respectively. The identification of Campylobacter at genus level from the extracted DNA of fecal specimen was done based on the size of amplicons on agarose 1% gel (Gene Fanavaran, Iran) in horizontal electrophoresis system (Padide Nozhen Pars, Iran) and gel documentation (Quantum ST4. Germany). Then, PCR products were sent to Macrogen company (South Korean) for sequencing and final confirmation by ABI Applied Biosystems 3730xl instrument.
Table 1.
Properties of primers used to detect of Campylobacter genus.
| Primers ID | Sequence (5' to 3') | Target gene | Size(bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| slyD-F | GCGAAGGTGAAAATGGCGAA | slyD | 216 | 62 |
| slyD-R | GATGATCGTGTCCATGTCCG | |||
| MD16S-F | ATCTAATGGCTTAACCATTAAAC | 16SrRNA | 857 | 56.4 |
| MD16S-R | GGACGGTAACTAGTTTAGTATT | |||
| cadF-F | TAAAAGCGGTGGATTTGGAC | cadF | 218 | 54 |
| cadF-R | CAGGACATTTTGCTTGTGGA | |||
| rpoA-F | CGAGCTTGCTTTGATGAGTG | rpoA | 121 | 52 |
| rpoA-R | AGTTCCCACAGGAAAACCTA | |||
| dnaJ-F | GGCAGGGGACAAGTAGGAA | dnaJ | 227 | 52 |
| dnaJ-R | CCCCTATTGCCACTTTTGCT |
Statistical Analysis
Statistical analyses were carried out using MedCal 18.11. P values less of 0.05 were considered as statistically significant.
Results
PCR results of housekeeping genes
Among of 60 samples, 21 samples (35%), 21 samples (35%), 20 samples (33.33%), samples (33.33%), 20 samples (33.33%), and 20 samples (33.33%) were positive for Campylobacter by slyD, cadF, dnaJ, rpoA , and 16SrRNA, respectively (Table 2).
Table 2.
Results of amplifications for each group.
| Genes | 16SrRNA | slyD | cadF | rpoA | dnaJ |
|---|---|---|---|---|---|
| Group | Positive number (Percentage) | ||||
| Healthy | 5 (16.66%) | 5 (16.66%) | 5 (16.66%) | 5 (16.66%) | 5 (16.66%) |
| Patient | 15(50%) | 16 (53%) | 16 (53%) | 15 (50%) | 15 (50%) |
Sequencing
Results of sequencing were analyzed by using Mega4 and Chromas software. (Figure 1) (a-e) showed amplifications and sequencing results of different genes in present study. Data were compared with sequences of gene bank and confirmed statistically significant using BLAST software.
Fig. 1.
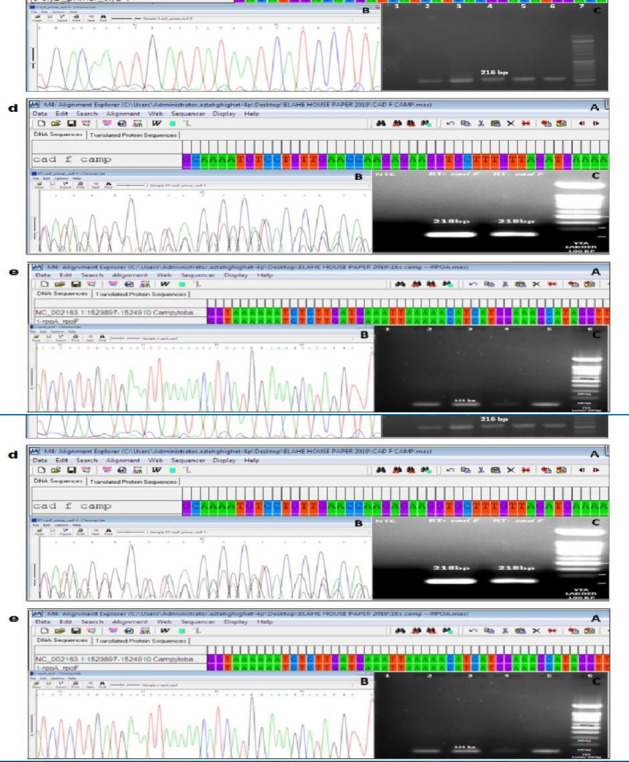
(A, B) a-e: Sequencing results in Mega4 and chromes software for 16SrRNA {max score: 67.1, QC: 40%, total score: 67.1,Percent Identity: 82.54 %, E value: 1e-09)}, dnaJ {max score: 301, QC: 92%, total score: 301, Percent Identity: 94.09%, E value: 5e-80}, slyD {max score: 234, QC: 79%, total score: 270, Percent Identity: 87.13%, E value: 7e-60}, cadF {max score: 301, QC: 92 %, total score: 301, Percent Identity: 94.09%, E value: 5e-80} and rpoA {max score: 115, QC: 76%, total score: 115, Percent Identity: 97.10%, E value: 4e-2}, (C) a-e: Results of PCR product for 16SrRNA, dnaJ, slyD, cadF and rpoA on 1.3% agarose gels. Product size: 857, 227, 216, 218 and 121bp.
Discussion
The increasing isolation of Campylobacter from clinical specimens and healthy people has increased its importance in public health (13). Due to the presence of Campylobacter in food products with the animal origin, vegetables, and water, this bacterium has been identified as a foodborn pathogen (14). In addition, Campylobacter can cause gastrointestinal diseases in humans (gastroenteritis) and abortions in animals (15). Since molecular methods are faster and more accurate than microbial culture methods, in this study we compared molecular diagnostic assay using different genes to detect Campylobacter in human fecal samples.
In previous studies, 16SrRNA gene was mainly used for molecular detection of Campylobacter species. Due to the existence of different types of bacterial genomes in the excreted DNA from the feces as well as repeated nucleotide sequences in the 16SrRNA gene in most bacteria, the identification of the Campylobacter is not sufficiently certain by using only 16SrRNA gene. Regarding the necessity of accurate detection of Campylobacter, this study examined the detection of these bacteria by using cadF, rpoA, dnaJ , and slyD genes compared with 16SrRNA gene. Bang et al. in 2003 proposed cadF, flaA , and ceuE genes for determination of Campylobacter by PCR assay(16). Ritz et al. in 2009 suggested usage of rpoA gene for genotyping of Campylobacter species by Multilocus sequence typing (MLST) and evaluation of each gene by using the real time-PCR assay(6). Konkel ME et al. in 1999 used different genes to detectCampylobacter, and used cadF gene for reproduction and sequencing of gene, and according to this study, this gene can be very suitable for diagnosis of Campylobacter (17). Datta et al. in 2003 determinated Campylobacter spp. with 11 virulence gene by multiplex-PCR assay. Among 11 primers designed for virulence genes, dnaJ, cadF, and cdtB were present in all samples were positive for Campylobacter (18).
In this study, not only 16SrRNA gene was used but also rpoA, cadF, slyD , and dnaJ genes were used as housekeeping genes for molecular detection of Campylobacter. Among of 30 samples of patients with diarrhea, 15 samples (50%) were positive for Campylobacter by 16SrRNA gene which was the same with the results of rpoA and dnaJ genes, While the results of slyD and cadF genes in patients with diarrhea were 16/30 (53.33%), therefor, the results of these genes were more sensitivity than 16SrRNA, dnaJ , and rpoA genes. Also among of healthy people samples, the results of all genes were consistent with each other, and among 30 healthy people samples, Campylobacter were identified in five healthy people samples by studied housekeeping genes. These data showed that cadF and slyD are probably the most suitable genes for accurate detection for Campylobacter. According to similar studies, cadF and cdtB have been identified as suitable genes for the detection this bacterium, and results of this gene are consistent with other studies. The dnaJ gene has been reported to be suitable for the diagnosis for Campylobacter and its results are consistent with the 16SrRNA gene. The rpoA gene in this study was able to detect 90% of Campylobacter isolates, and this result is consistent with other studies. Here, total DNA was extracted from the stool so it is a mixture of genome of different organisms and can the inconsistency with other studies. As shown in (Table 3) , the sensitivity and specificity of using cadF and slyD gene for detection of Campylobacter by PCR were 53.33% and 83.33%, respectively. The sensitivity and specificity of using these genes have been reported more than 16SrRNA, rpoA, and dnaJ housekeeping genes. Therefore, the amplification of the slyD and cadF genes with specific primers, can be adequate for identification of Campylobacter in both groups,patients with diarrhea and healthy people.
Table 3.
Sensitivity and specificity of PCR.
| Total Sample | Gene Diagnostic | Result | Person | Percentage | Diagnostic analysis | Chi-square analysis | p value | DF |
|---|---|---|---|---|---|---|---|---|
| 30 samples Patient + 30 samples healthy people | Detection With 16SrRNAgene | 20 positive | 15 Patient | 25% | Sensitivity: 50.00% | 7.375 | 0.006 | 1 |
| 5 healthy | 8.3% | (31.297%-68.703%) Specificity: 83.33% | ||||||
| 40 Negative | 15 Patient | 25% | (65.279%-94.358%) | |||||
| 25 healthy | 41.7% | Positive Predictive Value: 75.00% | ||||||
| (55.529%-87.816%) Negative Predictive Value: | ||||||||
| 62.50%(52.967%-71.153%) Disease prevalence: | ||||||||
| 50.00%(36.806%-63.194%) | ||||||||
| 30 samples Patient + 30 samples healthy people | Detection With cadF gene | 21 positive | 16 Patient | 26.7% | Sensitivity: 53.33% | 8.717 | 0.0032 | 1 |
| 5 healthy | 8.3% | (34.326%-71.658%) Specificity: 83.33% | ||||||
| 39 Negative | 14 Patient | 23.3% | (65.279%-94.358%) | |||||
| 25 healthy | 41.7% | Positive Predictive Value: 76.190% | ||||||
| (57.341%-88.396%) Negative Predictive Value: | ||||||||
| 64.103%(54.119%-72.998%) | ||||||||
| Disease prevalence: 50.00% | ||||||||
| (36.806%-63.194%) | ||||||||
| 30 samples Patient + 30 samples healthy people | Detection With slyDgene | 21 positive | 16 Patient | 26.7% | Sensitivity: 53.33% | 8.717 | 0.0032 | 1 |
| 5 healthy | 8.3% | (34.326%-71.658%) Specificity: 83.33% | ||||||
| 39 Negative | 14 Patient | 23.3% | >(65.279%-94.358%) | |||||
| 25 healthy | 41.7% | Positive Predictive Value: 76.190% | ||||||
| (57.341%-88.396%) Negative | ||||||||
| Predictive Value: 64.103% (54.119%-72.998%) | ||||||||
| Disease prevalence: 50.00% | ||||||||
| (36.806%-63.194%) | ||||||||
| 30 samples Patient + 30 samples healthy people | Detection With rpoA gene | 20 positive | 15 Patient | 25% | Sensitivity: 50.00% | 7.375 | 0.006 | 1 |
| 5 healthy | 8.3% | (31.297%-68.703%) Specificity: 83.33% | ||||||
| 40 Negative | 15 Patient | 25% | (65.279%-94.358%) | |||||
| 25 healthy | 41.7% | Positive Predictive Value: 75.00% | ||||||
| (55.529%-87.816%) Negative | ||||||||
| Predictive Value: 62.50% (52.967%-71.153%) | ||||||||
| Disease prevalence: 50.00% (36.806%-63.194%) | ||||||||
| 30 samples Patient + 30 samples healthy people | Detection With dnaJ gene | 20 positive | 15 Patient | 25% | 7.375 | 0.006 | 1 | |
| 5 healthy | 8.3% | Sensitivity: 50.00% | ||||||
| (31.297%-68.703%) Specificity: 83.33% | ||||||||
| (65.279%-94.358%) | ||||||||
| 40 Negative | 15 Patient | 25% | Positive Predictive Value: 75.00% | |||||
| (55.529%-87.816%) Negative | ||||||||
| 25 healthy | 41.7% | Predictive Value: 62.50% (52.967%-71.153%) | ||||||
| Disease prevalence: 50.00% (36.806%-63.194%) |
In this study, we showed that Campylobacter can be detected from a mixture of different genomes with the highest accuracy using housekeeping genes, including cadF, slyD, dnaJ , and rpoA, without using the 16srRNA gene. Probably, replication of cadF and slyD genes can be considered as a alternative for 16SrRNA in diagnosis of Campylobacter, however research in a larger statistical society is suggested.
The application of PCR molecular method is a more accurate method for diagnosis of Campylobacter bacteria in genomic DNA extracted from the feces. The diagnostic standard in different laboratories is based on the usage of the 16SrRNA gene. One of important implications of our study is molecular detection of campylobacter within template DNA that has been purified from the stool with mixture of genomes of different organisms. In this study, we showed that campylobacter can be detected from a mixture of different genomes with the highest accuracy using housekeeping genes, including cadF, slyD, dnaJ and rpoA, Apart from the 16srRNA gene. Probably,replication of cadF and slyD genes can be considered as an alternative for 16SrRNA in molecular detection of Campylobacter.Campylobacter species.
Acknowledgement
This study is performed in Arak University of Medical Sciences. We are thankful to the staff of Infectious Diseases Research Center of Arak University of Medical Sciences for her technical assistance.
The authors declare they have no conflict of interests.
References
- 1.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, et al. Campylobacter spp. as a foodborne pathogen: a review. Front Microbiol. 2011;2(200) doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuijten PJ, Bartels C, Bleumink-Pluym NM, Gaastra W, van der Zeijst BA. Size and physical map of the Campylobacter jejuni chromosome. Nucleic Acids Res. 1990;18(21):6211–6214. doi: 10.1093/nar/18.21.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. Campylobacter jejuni—an emerging foodborne pathogen. Emerg Infect Dis. 1999;5(1):28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassenaar TM. Toxin production by Campylobacter spp. Clin Microbiol Rev. 1997;10(3):446–476. doi: 10.1128/cmr.10.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laprade N, Cloutier M, Lapen DR, Topp E, Wilkes G, Villemur R, et al. Detection of virulence, antibiotic resistance and toxin (VAT) genes in Campylobacter species using newly developed multiplex PCR assays. J Microbiol Methods. 2016;124:41–7. doi: 10.1016/j.mimet.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Ritz M, Garenaux A, Berge M, Federighi M. Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RTqPCR and application to oxidative stress. . J Microbiol Methods. 2009;76(2):196–200. doi: 10.1016/j.mimet.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J bacteriol. 1998;180(18):4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345(6270 ):63–5. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 10.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29(1):181–4. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186(9):2629–35. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Case RJ, Boucher Y, Dahllöf I, Holmström C, Doolittle WF, Kjelleberg S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007;73(1):278–88. doi: 10.1128/AEM.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leblanc-Maridor M, Garénaux A, Beaudeau F, Chidaine B, Seegers H, Denis M, et al. Quantification of Campylobacter spp. in pig feces by direct real-time PCR with an internal control of extraction and amplification. . J Microbiol Methods. 2011;85(1):53–61. doi: 10.1016/j.mimet.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Kang CR, Bang JH, Cho S-I. Campylobacter jejuni Foodborne Infection Associated with Crosscontamination: Outbreak in Seoul in 2017. Infect Chemother. 2019;51(1):21–27. doi: 10.3947/ic.2019.51.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolton DJ. Campylobacter virulence and survival factors. Food microbiol. 2015;48:99–108. doi: 10.1016/j.fm.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Bang DD, Scheutz F, Oren Gradel K, Nielsen E, Pedersen K, Engberg J, et al. Detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from different sources and cytolethal distending toxin production suggest potential diversity of pathogenic properties among isolates. Genome Letters. 2003;2:62–72. [Google Scholar]
- 17.Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Identification of proteins required for the internalization of Campylobacter jejuni intocultured mammalian cells. Adv Exp Med Biol. 1999;473:215–24. doi: 10.1007/978-1-4615-4143-1_22. [DOI] [PubMed] [Google Scholar]
- 18.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003;52(pt 4):345–348. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]


