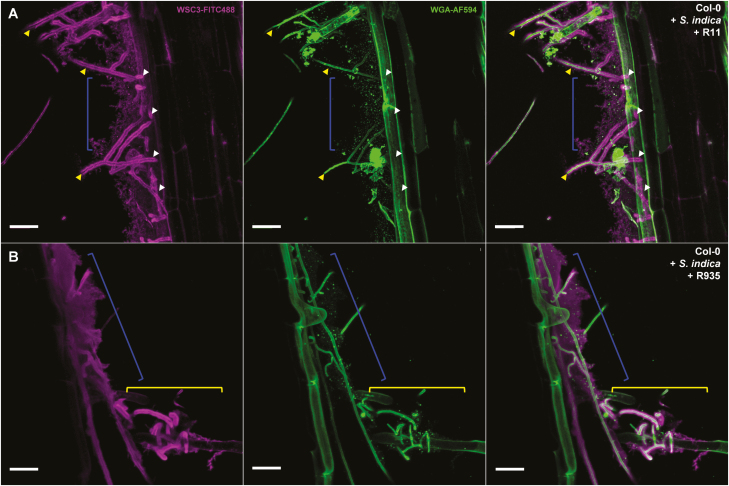
Fig. 2.
Live cell images of root-associated bacteria embedded in the fungal β-glucan matrix. Confocal laser scanning microscopy live stain images of an A. thaliana Col-0 root segment, 12 d post-colonization with the fungus Serendipita indica and root-associated bacteria R11 (Bacillus sp.) (A) or R935 (Flavobacterium sp.) (B) (Bai et al., 2015). The fungal cell wall and bacteria (dots) were stainable with WGA-AF594 (green pseudo-color). β-1,3-glucan was visualized using the FITC488-labeled lectin WSC3 (magenta pseudo-color) (Wawra et al., 2019). Fungal chitin was only detected in hyphae growing in the extracellular space (yellow arrowheads), whereas the β-glucan matrix was also detectable after the hyphae entered the root cortical cells (white arrowheads). Interestingly, the β-glucan architecture was very distinct in the presence of the individual bacterial strains (structures indicated by the blue brackets). While a fine structure in the glucan network was visible for S. indica in combination with R11, the combination with R935 resulted in an apparently extended, more dense, and amorphous looking β-glucan layer. In addition, in the presence of R935, the fungal β-glucan matrix around the S. indica hyphae appeared more diffuse and less compact compared with the matrix detected around more isolated growing hyphae (B, yellow brackets). It is currently unclear whether the additionally deposited β-glucan is produced by the bacteria themselves or whether their presence triggers enhanced production and secretion by the fungus. Scale bars=25 µm.