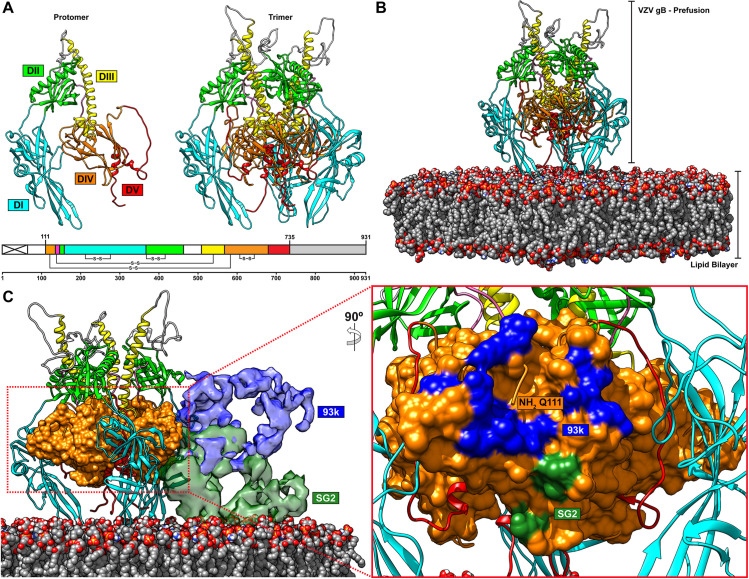
Fig 11. The neutralizing epitope of mAb 93k is accessible on the prefusion form of VZV gB.
A–A homology model of VZV gB in a prefusion conformation based on the 9.0Å cryo-EM structure of HSV gB [37]. The left-hand panel shows a single VZV gB protomer and the right-hand panel is the complete gB trimer in the prefusion conformation. The linear structure of VZV gB below the two panels depicts the colors used for the homology model; the signal sequence (crossed white box), DI (cyan), DII (green), DIII (yellow), DIV (orange), DV (red), linker region (hot pink) and CTD (grey box). B and C–Snapshots taken from S4 Movie. B–The prefusion conformation of VZV gB modelled in conext with a lipid bilayer composed of lipids SM (3.1%), PC (51.2%), PI (11.3%), PS (4.6%), PE (29.8%) found in herpesvirus virions [85]. C–The mAb 93k (blue) and SG2 (green) Fabs bound to the prefusion conformation of VZV gB. Domian IV (orange) of VZV gB is represented in surface. The presence of the lipid bilayer does not prevent accesibility of the 93k epitope but will prevent SG2 from binding to gB in the prefusion conformation. The left-hand panel shows a rotated (90°) and zoomed in region of gB DIV and the footpring of the 93k and SG2 Fabs as determined in Fig 3.