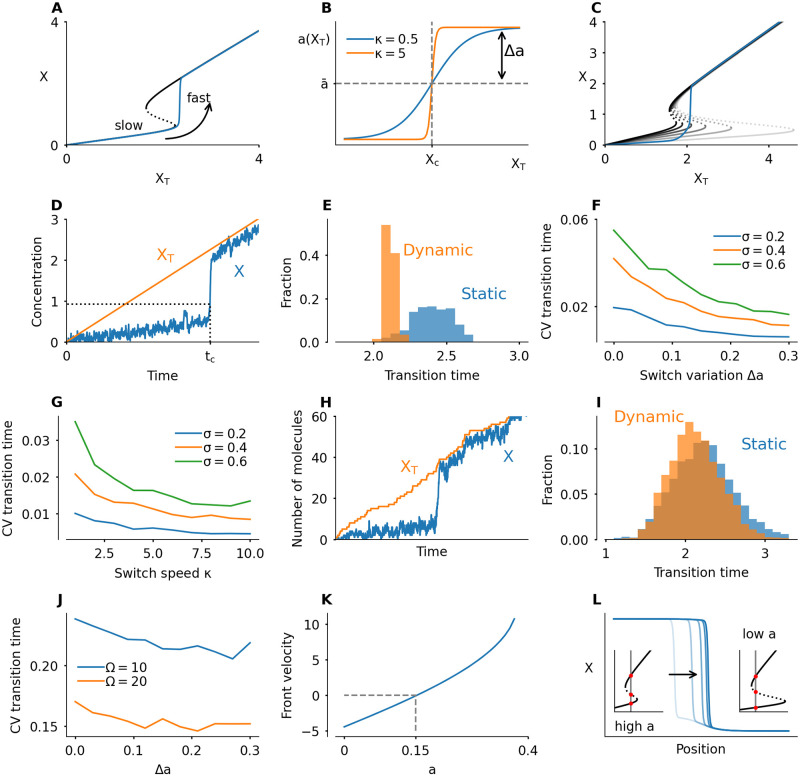
Fig 4. A dynamically changing bistable switch enhances robustness and accuracy of transitions in time and space.
A) When XT increases at a constant rate (here kX = 0.2), the activity of the protein will increase suddenly at the moment XT crosses the activation threshold of the switch. B) Function used to make the shape of the switch dependent on the total amount of protein, by coupling a to XT. The switch variation depends on the value of Δa, which controls the magnitude of possible deviations of a from a mean value . The parameter κ controls how abruptly the system switches between low and high a values. C) Time evolution of a system in which XT increases at a constant rate, and a is coupled to XT. The gray response curves are snapshots in time. The activation threshold starts out to the far right, and moves left as XT, and with it a, increases. Here . D) Evolution of a system in which noise is added to the X variable (Langevin version). The transition time tc is defined as the time when X crosses a threshold value. Here . E) Histogram of measured transition times for a static switch (Δa = 0) and a dynamic switch (Δa = 0.3), with . The spread is lower for the dynamic switch. F) Coefficient of variation (CV), defined as standard deviation divided by mean, of the transition time, as function of the switch variation Δa. Here . G) Coefficient of variation as function of κ, which defines the speed by which a changes. Faster changing corresponds to smaller deviations. Here . H) Time series of a run of the Gillespie algorithm/stochastic simulation algorithm (SSA). Parameter values are the same as in panel D, but here molecule counts are discrete. We used a system size Ω = 20. I) Histogram of transition times in the stochastic simulation algorithm (compare to panel E) with Ω = 20. The variation is lower for the dynamic switch, but the difference is much smaller than in the Langevin version. J) Coefficient of variation as function of Δa for the stochastic simulation algorithm (compare to Panel F). K) Velocity of a bistable front as function of a, for XT = 2. A positive velocity means that the active protein state overtakes the inactive state (the front shown in panel L moves to the right). At a ≈ 0.15, the front is stationary. L) A bistable front in the presence of an inhomogeneous a profile in space. On the left, a = 0.27, which means the front moves to the right. On the far right, a = 0.13 is low, which means that the front moves to the left. The result is that the front is pinned in the middle where a ≈ 0.15. This pinning can be lifted by a redistribution of a (see S2 Video). Other parameters for all panels can be found in the Methods section, Table 1. Histograms and coefficients of variations where calculated over 200 simulations for the Langevin version (panels E to G) and 2000 simulations for the SSA (H to J).