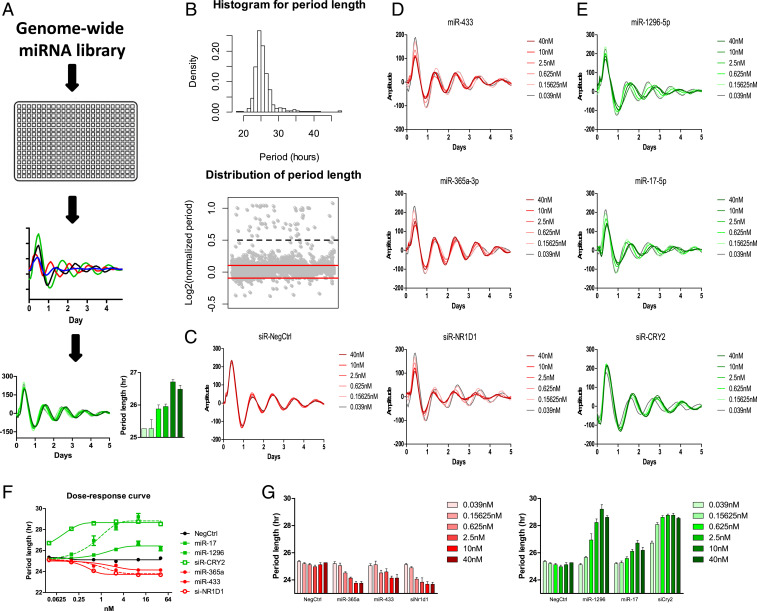
Fig. 1.
A cell-based genome-wide screen for miRNA modifiers of circadian rhythms. (A) A schematic diagram of the genome-wide miRNA screen. Bmal1-dLuc and Per2-dLuc reporter cells were transfected with miRNAs in 384-well plates. Bioluminescence was recorded and analyzed to obtain circadian parameters and select candidate hits. Validation was performed by measuring the dose-dependent effects of some candidate miRNAs on circadian phenotypes. (B) Distribution of circadian period lengths of the screen. The histogram shows a tail toward the long period. Gray dots in the bottom panel represent normalized period lengths. The period length of each well was divided by the negative control and indicated in Log2 space. The cutoff was −0.1 and +0.1 (red lines). Values above 0.5 (black dash line) were considered outliers. (C) Representative bioluminescence profiles for dose-dependent phenotypic validation of negative control, short-period positive control (siNR1D1), and long-period positive control (siCRY2). (D) Short-period candidate hits (miR-433, miR-365a-3p) and (E) long-period candidate hits (miR-1296-5p, miR-17). (F) The dose–response curve of the indicated candidate miRNAs and control RNAs. (G) The period length parameters of the indicated candidate miRNAs and control RNAs. Per2-dLuc U2OS cells were transfected with the indicated amount of miRNAs (six 4-fold dilution series from 0.039 to 40 nM/well). Data represent the mean ± SD (n = 4).