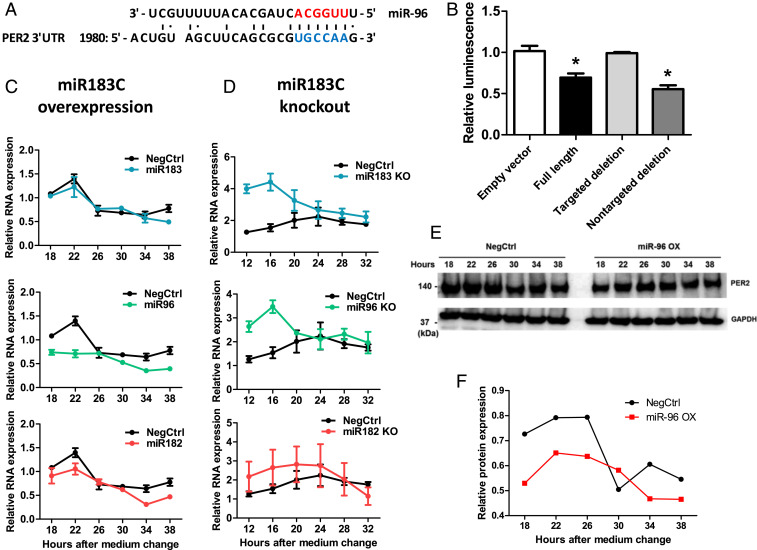
Fig. 5.
PER2 was a direct target of miR-96. (A) Sequence alignment between miR-96 and its putative binding sites (blue letters) in the PER2-3′ UTR. Red letters indicate canonical seed nucleotides. (B) HEK-293T cells were transfected with a luciferase reporter plasmid containing one of the following sequences: 1) no 3′ UTR; 2) a full-length wild-type PER2-3′ UTR; 3) a full-length PER2-3′ UTR with a targeted deletion at the predicted binding site of miR-96 (1992_2002 del: gcgcgTGCCAA; uppercase is the potential seed-binding site of miR-96); 4) a full-length PER2-3′ UTR with a deletion in a sequence (1712_1734 del: ttcataaacacaagaacacttta) predicted not to be bound by miR-96, *P < 0.05. (C) PER2 mRNA levels in U2OS cells that transfected with miRNA mimics of each member of the miR-183/96/182 cluster. RNA was sampled 18 h after medium change, every 4 h across 1 d. The mRNA levels were determined by qPCR (n = 3). (D) PER2 mRNA levels in miR-183/96/182 cluster knockout cells. RNA from U2OS cells with the deletion of each member of the miR-183/96/182 cluster was harvested 12 h after medium change, every 4 h across 1 d. The mRNA levels were determined by qPCR (n = 3). Data represent the mean ± SEM (E) U2OS cells were transfected with either control or miR-96 mimics, and PER2 protein levels were analyzed by Western blot 48 h posttransfection. (F) Quantification of PER2 protein band intensity compared with the negative control on Western blot.