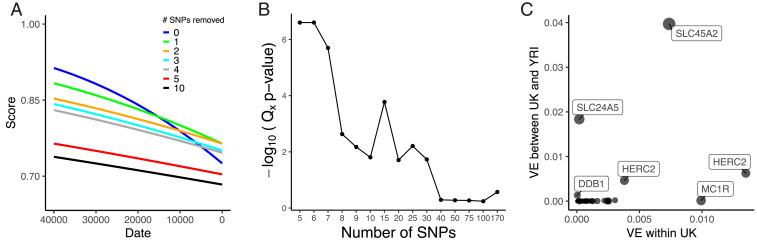
Fig. 4.
(A) Regressions of genetic score based on UK Biobank SNPs using the capture-shotgun dataset over date, with scores using all SNPs and iteratively removing top GWAS effect size SNPs. (B) Qx empirical −log10(P values) using UK Biobank skin pigmentation-associated variants with all 1000 Genomes populations. Different numbers of SNPs were used to calculate Qx, which were ordered by GWAS-estimated effect size. (C) Variance explained by UK Biobank SNPs with nearest gene labeled between UK and YRI populations (y axis) and within the UK population (x axis).