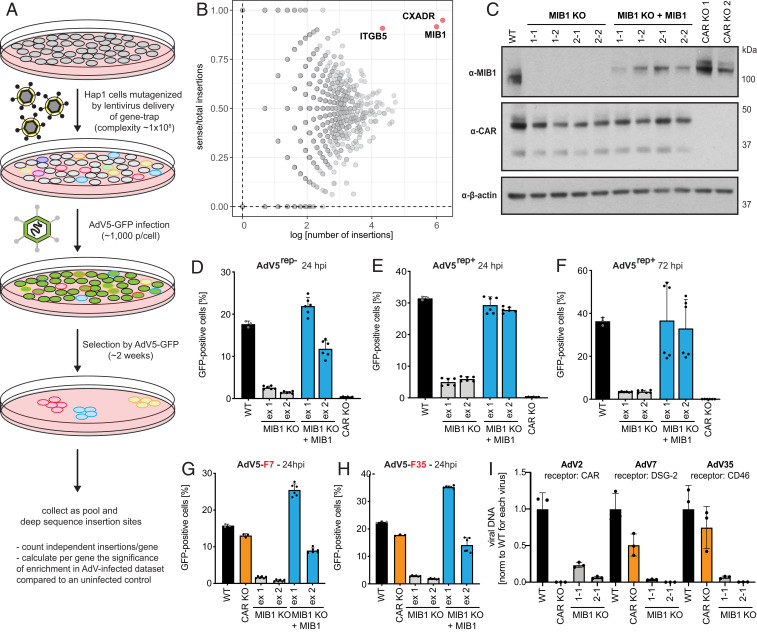
Fig. 1.
Haploid genetic screen identifies E3 ubiquitin ligase MIB1 as a host factor necessary for AdV infection independent of AdV receptor. (A) Outline of haploid genetic screen for AdV5 infection. (B) Fishtail plot showing enrichment of genes in haploid screen upon AdV5 selection identified by deep sequencing and plotted as number of distinct gene-trap insertions (x axis) and the ratio of sense over total insertions (y axis) per gene. Genes significantly enriched for sense orientation integrations (FDR < 0.05) labeled by gene name (red). (C) Western blot of MIB1 CRISPR KO clones in Hap1 cells generated by guide RNAs targeted to either exon 1 (1-1, 1-2) or exon 2 (2-1, 2-2), KO clones reconstituted with MIB1, and two CAR KO clones probed for MIB1, CAR (two isoforms), or β-actin expression. (D–F) Infection of Hap1 WT cells, MIB1 KO clones, WT MIB1-reconstituted KO clones, and CAR KO clones with (D) AdV5rep−-GFP (MOI = 2,500 p/cell; 24 hpi). (E and F) AdV5rep+-GFP [(E) MOI = 75 p/cell; 24 hpi and (F) MOI = 50 p/cell; 72 hpi] and (G and H) Infection of Hap1 WT cells, a CAR KO clone, MIB1 KO clones, and WT MIB1-reconstituted KO clones with recombinant AdV5rep−-GFP expressing the fiber protein of (G) AdV7 or (H) AdV35 (MOI = 1 × 104 p/cell; 24 hpi). (I) Hap1 WT cells, a CAR KO clone, and individual MIB1 KO clones (1-1, 2-1), infected with WT AdV serotypes using distinct entry receptors: AdV2 (CAR), AdV7 (DSG-2), and AdV35 (CD46) at MOI = 1 × 103 p/cell (vDNA was harvested at 72 hpi and measured by qPCR). All experiments were performed in triplicate and, if not indicated otherwise, harvested cells were analyzed by flow cytometry and plotted as the percentage of GFP-positive cells averaged across the two selected MIB1 KO clones for guide RNAs targeting either exon 1 (ex 1) or exon 2 (ex 2).