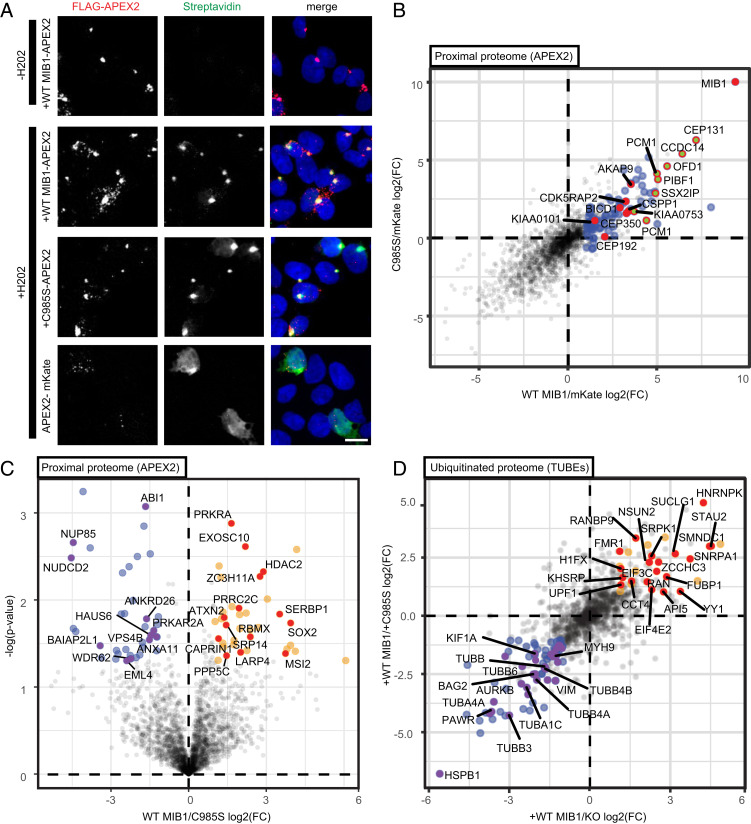
Fig. 4.
Proteomic approaches confirm MIB1 localization to centriolar satellites in AdV-infected Hap1 cells and reveal MIB1 ubiquitination-dependent association with RNPs. (A) Uninfected MIB1 KO clone 2-1 reconstituted with FLAG-APEX2–tagged WT MIB1, C985S MIB1, or mKate were incubated with biotin-phenol for 1 h and pulse labeled with H2O2 followed by fixation and staining with AF488-coupled streptavidin for biotinylated proteins (green), the indicated FLAG-APEX2–tagged construct (red: anti-FLAG), and DRAQ5 nuclear stain (blue). (Scale bar: 10 µm, maximum projected.) (B and C) For LC-MS/MS, MIB1 KO clone 2-1 reconstituted with FLAG-APEX2 fused to either mKate, WT MIB1, or C985S MIB1 were incubated with biotin-phenol and infected with AdV5rep+-GFP (MOI = 200p/cell), pulse-labeled with H2O2 at 70 min pi, and immediately collected for lysate preparation and streptavidin pulldown to isolate biotinylated proteins (n = 3). (B) Proximal protein enrichment (FC) over APEX2-mKate is plotted as log2 of the ratio of protein abundance for WT MIB1/mKate (x axis) and C985S MIB1/mKate (y axis). GO terms analysis of proteins twofold or greater (P value <0.05) in the WT MIB1/mKate comparison (blue) shows enrichment of centriolar satellite proteins (green, red rim), as a subset of centrosome proteins (red). (C) Ubiquitination-dependent proximal proteome determined by direct comparison of APEX2-tagged WT MIB1 and C985S MIB1 (unique peptides ≥1) plotted as log2 of the ratio of protein abundance for WT MIB1/C985S MIB1 (x axis) and −log(P value) (y axis). GO terms analysis of those proteins enriched by twofold or greater (orange) or depleted by twofold or greater (blue) in proximity to WT MIB1 relative to C985S MIB1 (P value <0.05) reveals enrichment of RNA-binding proteins (red with gene names labeled) and depletion of cytoskeletal proteins (purple with gene names labeled). (D) Identification of differentially ubiquitinated proteins between MIB1 KO clone 2-1 and KO reconstituted with either WT MIB1 or C985S MIB1 infected with AdV5rep+-GFP (MOI = 200 p/cell) (n = 3). At 1 hpi, cells were harvested and endogenous ubiquitinated proteins were isolated using TUBE-Dynabeads followed by LC-MS/MS and plotted as log2 of the ratio of protein abundance for WT MIB1/MIB1 KO (x axis) and WT MIB1/C985S MIB1 (y axis). GO terms analysis of those proteins enriched twofold or greater in both WT MIB1/KO and WT MIB1/C985S (P value <0.05 in either) (orange) or depleted by twofold or greater in WT MIB1/KO and WT MIB1/C985S (P value <0.05 in either) (blue) reflect RNA binding (red with all gene names labeled) or cytoskeleton (purple with select proteins significant to both WT MIB1/KO and WT MIB1/C985S condition labeled by gene name), respectively.