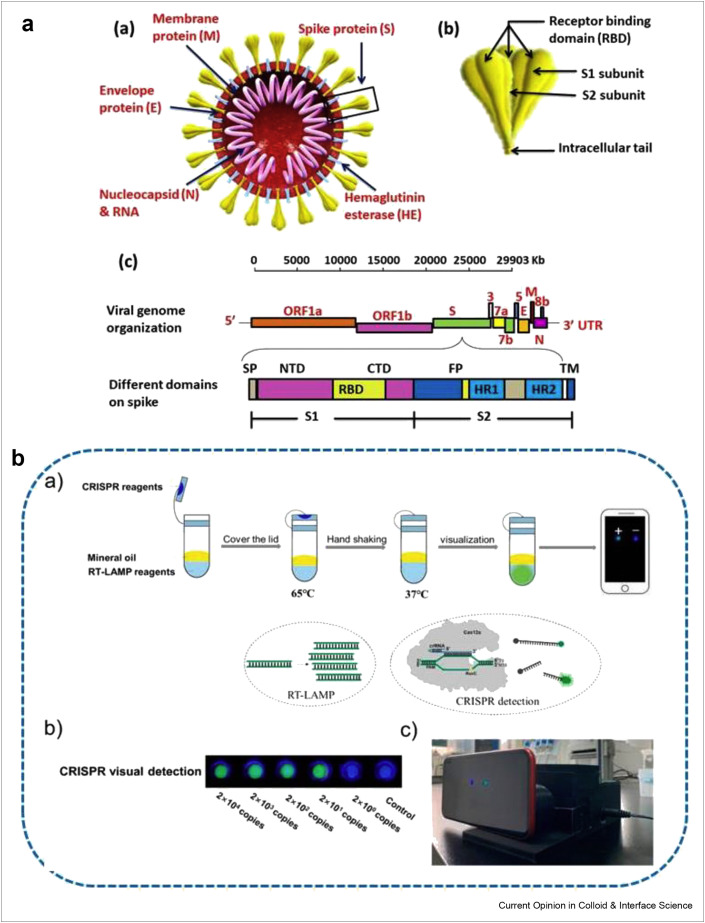
Figure 1.
The detailed genomic representation of SARS-CoV-2 and its mobile phone detection (a) Structural illustration of SARS-CoV-2 (a) structural image of SARS-CoV-2, containing four different types of structural proteins (b) is the inset of key features of spike protein, representing the receptor binding domain, S1 and S2 regions, whereas (c) shows the full-length viral genome (29,903 nucleotides long) single-stranded positive RNA sequence. The leader sequence at the 5′ end and poly A tail ends at the 3′, contains structural proteins E,M,N,S along with different accessory genes (ORF 1a, 1b, 3a, 6, 7a, 7b, and 8b). The open reading frame ORF1a/b ‘the replicase’ is responsible for the coding of polymerase enzymes for viral RNA synthesis as well as non-structural proteins. The S1 subunit of spike protein consist of N-terminal domain (NTD), receptor binding domain (RBD) and C-terminal domain, whereas the S2 subunit comprises heptad repeat 1and 2, which plays a critical role in viral entry (b) The flow chart diagram describing the whole experimental process and principle for contamination-free visual detection method to detect different concentrations of RNA. Reprinted with permission from Ref. [56].