Abstract
Objective
To evaluate the association of MRI features with the major genomic profiles and prognosis of World Health Organization grade III (G3) gliomas compared with those of glioblastomas (GBMs).
Materials and Methods
We enrolled 76 G3 glioma and 155 GBM patients with pathologically confirmed disease who had pretreatment brain MRI and major genetic information of tumors. Qualitative and quantitative imaging features, including volumetrics and histogram parameters, such as normalized cerebral blood volume (nCBV), cerebral blood flow (nCBF), and apparent diffusion coefficient (nADC) were evaluated. The G3 gliomas were divided into three groups for the analysis: with this isocitrate dehydrogenase (IDH)-mutation, IDH mutation and a chromosome arm 1p/19q-codeleted (IDHmut1p/19qdel), IDH mutation, 1p/19q-nondeleted (IDHmut1p/19qnondel), and IDH wildtype (IDHwt). A prediction model for the genetic profiles of G3 gliomas was developed and validated on a separate cohort. Both the quantitative and qualitative imaging parameters and progression-free survival (PFS) of G3 gliomas were compared and survival analysis was performed. Moreover, the imaging parameters and PFS between IDHwt G3 gliomas and GBMs were compared.
Results
IDHmut G3 gliomas showed a larger volume (p = 0.017), lower nCBF (p = 0.048), and higher nADC (p = 0.007) than IDHwt. Between the IDHmut tumors, IDHmut1p/19qdel G3 gliomas had higher nCBV (p = 0.024) and lower nADC (p = 0.002) than IDHmut1p/19qnondel G3 gliomas. Moreover, IDHmut1p/19qdel tumors had the best prognosis and IDHwt tumors had the worst prognosis among G3 gliomas (p < 0.001). PFS was significantly associated with the 95th percentile values of nCBV and nCBF in G3 gliomas. There was no significant difference in neither PFS nor imaging features between IDHwt G3 gliomas and IDHwt GBMs.
Conclusion
We found significant differences in MRI features, including volumetrics, CBV, and ADC, in G3 gliomas, according to IDH mutation and 1p/19q codeletion status, which can be utilized for the prediction of genomic profiles and the prognosis of G3 glioma patients. The MRI signatures and prognosis of IDHwt G3 gliomas tend to follow those of IDHwt GBMs.
Keywords: Magnetic resonance imaging, Glioma, Glioblastoma, Isocitrate dehydrogenase, Apparent diffusion coefficient, Cerebral blood volume, Cerebral blood flow
INTRODUCTION
Diffuse lower grade gliomas (LGGs) are infiltrative brain neoplasms that conventionally include World Health Organization (WHO) grade II (G2) and III (G3) astrocytomas and oligodendrogliomas based on histological characteristics (1). These tumors demonstrate high variability in prognosis, with varied survival that ranges from 1 to 15 years (2). The most recent WHO classification has incorporated the genomic characteristics of these tumors with conventional phenotypic characteristics to produce an integrated diagnosis for diffuse gliomas, as recent studies regarding genomic analysis of gliomas have provided a better understanding of LGGs (1).
The two most well-known genetic mutations incorporated in the newest WHO guidelines are an isocitrate dehydrogenase (IDH)-1 mutation and a chromosome arm 1p/19q (1p/19q) codeletion. The tumors with this IDH mutation (IDHmut) have a better prognosis with a higher sensitivity to chemoradiation therapy than those without this IDH wildtype (IDHwt) (3,4). Codeletion of 1p/19q is another genetic modification that is associated with a favorable prognosis and treatment response (5,6) and is commonly found in oligodendroglial tumors.
Magnetic resonance imaging (MRI) is the most reliable imaging modality for the brain and provides not only geographic spatial characteristics, but also functional and molecular information about brain tissue by using various updated techniques, such as perfusion- and diffusion-weighted images. The correlation of genomic information with imaging parameters to identify the characteristics of G2 and G3 gliomas has been investigated in previous studies (7,8,9,10,11). Even with these interesting findings, the association of genetic profiles with imaging parameters of G3 gliomas still remains somewhat controversial. Even though one previous study has analyzed VASARI features of G3 anaplastic gliomas (12), there is no previous study that includes a large number of patients to fully elucidate both the qualitative and quantitative MR characteristics of G3 gliomas. Therefore, we found the need for a comprehensive investigation of the quantitative and qualitative imaging parameters of G3 gliomas to identify the specific imaging parameters that distinguish G3 glioma subtypes according to their genetic profiles. Moreover, as the IDHwt LGGs are known to behave similarly to IDHwt glioblastomas (GBMs) (7), comparing the imaging findings and prognosis of IDHwt G3 gliomas and GBMs can provide advantages in clinical therapeutic management planning and prognosis prediction for glioma patients.
Thus, the purpose of our study is to investigate the association of the qualitative and quantitative MRI features and major genomic profiles of G3 gliomas and to compare the imaging findings and prognosis between IDHwt gliomas and GBMs.
MATERIALS AND METHODS
Patients
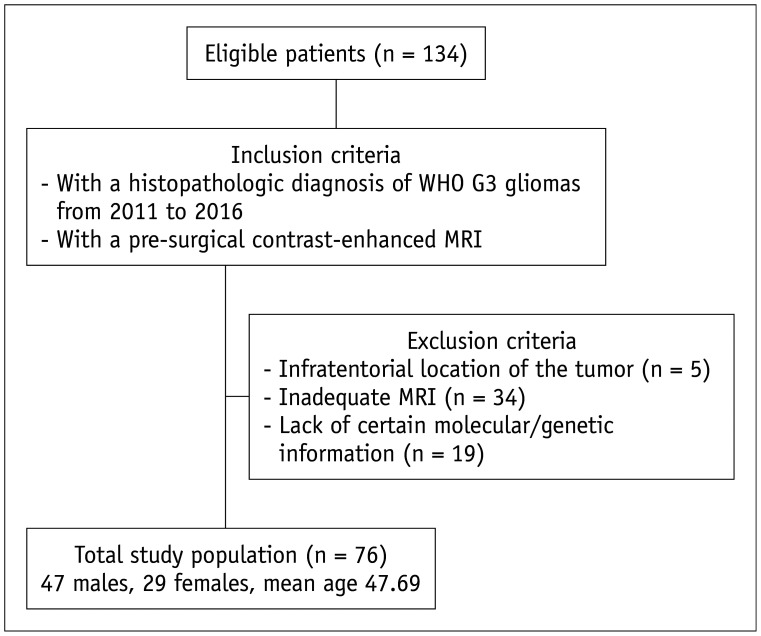
This retrospective study was approved by the Institutional Review Board of Seoul National University Hospital. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The requirement for informed consent was waived. One hundred thirty-four patients who were initially diagnosed with astrocytomas and oligodendrogliomas between January 2011 and September 2016 at our institution were consecutively included in the study. Using the exclusion criteria, a total of 76 patients were included in our study (47 males, 29 females; mean age, 47.69 ± 14.86 years; age range, 19 to 68 years). A detailed flow diagram of patient selection and classification is shown in Figure 1.
Fig. 1. Flowchart for selection of the study population.
G3 = World Health Organization grade III
To compare the imaging features of IDHwt LGGs and GBMs, we utilized the same inclusion and exclusion criteria mentioned above for G3 gliomas, except a requirement of histopathologic diagnosis of GBM was used for the GBM study. A total of 155 IDHwt GBM patients were included in the comparison.
Image Acquisition
For each patient, MR images were acquired by using a 1.5- or 3T MRI scanner with an 8-, 32-, or 64-channel head coil. MR scan parameters are summarized in Supplementary Table 1.
Image Processing
The conventional MR images, apparent diffusion coefficient (ADC) maps, and dynamic susceptibility contrast-enhanced perfusion-weighted imaging were digitally transferred from the picture-archiving and communication system (Infinitt, Infinitt) workstation to a personal computer for further analysis. All imaging analyses were performed by using a dedicated software package (NordicICE, NordicNeuroLab).
Image Analysis
Location, presence of an enhancing portion, necrosis, T2 hypersignal intensity margin of the tumor, and presence of a “T2-fluid attenuated inversion recovery (FLAIR) mismatch sign” were evaluated (Supplementary Material 1). One investigator, (with 3 years in brain MRI experience), who was blinded to the patient information and data, performed all image analyses and drew polygonal regions of interest (ROIs) that contained the T2 high signal intensity lesions in each section of resliced T2-weighted image (T2WI) and FLAIR images, including the cystic and necrotic regions. All of the ROIs were confirmed by an expert, board-certified neuroradiologist, (with 15 years in brain MRI experience).
Detailed information of image analysis is depicted in Supplementary Material 1 and description of the imaging parameters is summarized in Supplementary Table 2.
Molecular/Genetic Analysis
Detailed explanation of molecular genetic analysis is demonstrated in Supplementary Material 2.
Progression-Free Survival
Progression-free survival (PFS) was calculated from the date of initial diagnosis and progression according to the Response Assessment in Neuro-Oncology criteria (13). We only considered the first instance of progression or death, whichever comes first, and all the G3 glioma patients included in the study underwent surgical resection or biopsy, followed by radiation therapy as routine treatment for G3 glioma at our institution.
Validation Data Set
We utilized another set of G3 glioma cases to validate our predictive imaging parameters derived from the multivariable analysis to identify any association between the imaging features of G3 gliomas and the genetic profiles of the tumors. From October 2016 to July 2017, 17 G3 glioma patients with preoperative MRI, histologic diagnoses, and genetic information were available for evaluation. The quantitative imaging parameters were evaluated using the same methodology as mentioned above.
Statistical Analysis
All statistical analyses were performed with MedCalc (version 12.1.0 for Microsoft Windows 2000/XP/Vista/7, MedCalc Software). The results with a p value of less than 0.05 were considered statistically significant.
First, a comparison of the imaging features between the IDHmut group and IDHwt group was performed, and subsequently, a comparison of the imaging features between IDH-mutant, 1p/19q-codeleted (IDHmut1p/19qdel) and IDH-mutant, 1p/19q-nondeleted (IDHmut1p/19qnondel) was performed. Variables were compared between the two groups using unpaired Student's t test, Mann-Whitney U test, and Fisher's exact test. Subsequently, univariable and multivariable regression analyses were used to determine significant associations between the imaging parameters and the molecular information of G3 gliomas.
Then, the G3 gliomas were divided into three groups according to the different genetic profiles for the comparison of imaging features using ANOVA: IDHmut1p/19qdel, IDHmut1p/19qnondel, and IDHwt. Additionally, using the Kaplan-Meier method, we compared the PFS according to the different genetic profiles of the tumors. Survival curves were compared using a log-rank test and multivariable analysis was performed using the Cox proportional hazards model to find correlation between imaging features and PFS.
In addition, comparison of imaging features was performed using unpaired Student's t test, Mann-Whitney U test, and Fisher's exact test, and comparison of survival between IDHwt G3 gliomas and IDHwt GBMs was performed using Kaplan-Meier method.
To evaluate the reproducibility of the quantitative analysis and interobserver agreement, we utilized the intraclass correlation coefficient (ICC). Detailed description of statistical analysis for interobserver agreement is depicted in Supplementary Material 3.
RESULTS
Patient Clinical Parameters and Histological and Molecular Characteristics of the Tumor
Table 1 summarizes the patient demographics, pathological diagnosis, and the genetic characteristics of the tumors included in the study.
Table 1. Patient Demographics and Genetic Profiles of the Tumors of 76 Patients.
| Parameter | No. of Tumors (Total = 76) |
|---|---|
| Sex (%) | |
| Male | 47 (61.8) |
| Female | 29 (38.1) |
| Mean age, years (range) | 47.69 (19–68) |
| Pathologic diagnosis (%) | |
| Anaplastic astrocytoma | 57 (75.0) |
| Anaplastic oligodendroglioma | 19 (25.0) |
| Mean follow up (in months) | 16.3 |
| Number of events (%) | 22 (28.9) |
| Genetic information (%) | |
| IDH mutation | |
| Mutant | 47 (61.8) |
| Wild type | 29 (38.1) |
| 1p/19q | |
| Codeletion | 19 (25.0) |
| No deletion | 57 (75.0) |
| ATRX | |
| Loss | 16 (42.1) |
| No loss | 22 (57.9) |
| NA | 40 (52.6) |
| MGMT promoter | |
| Methylated | 46 (60.5) |
| Unmethylated | 30 (39.5) |
| EGFR | |
| Amplified | 3 (4.1) |
| Non-amplified | 71 (95.9) |
| NA | 2 (2.6) |
EGFR = epidermal growth factor receptor, IDH = isocitrate dehydrogenase, MGMT = O6-alkylguanine DNA alkyltransferase, NA = not available, 1p/19q = chromosome arm 1p/19q
Qualitative Analysis
IDHmut tumors presented with higher proportions of tumors with necrosis than IDHwt tumors (p = 0.008). Among the IDHmut tumors, the IDHmut1p/19del group presented with a higher proportion of enhancement (p = 0.004) (Supplementary Table 3). The T2-FLAIR mismatch sign was present in 28 of the 76 (36.8%) G3 gliomas included in the study. The accuracy, sensitivity, and specificity of the T2-FLAIR mismatch sign for the prediction of IDHmut1p/19qnondel G3 gliomas were 81.6%, 64.3%, and 91.7%, respectively. There was no statistically significant difference in lobar involvement or the proportion of well-defined margins of tumors according to histologic classification or genetic features of the tumor.
Quantitative Analysis
The IDHmut tumors showed a larger tumor volume in the T2WI (p = 0.017) than the IDHwt tumors. Additionally, the IDHmut tumors had a lower cerebral blood flow (CBF)_mean and a higher ADC_mean than the IDHwt tumors (p = 0.048 and 0.007, respectively) (Table 2). Among the IDHmut gliomas, the IDHmut1p/19qdel group had a higher cerebral blood volume (CBV)_mean (p = 0.024) and CBV_85 (p = 0.039). In addition, the IDHmut1p/19qdel group demonstrated a lower ADC_mean, ADC_5, ADC_10, and ADC_15 than the IDHmut1p/19qnondel group (all p values < 0.050) (Table 3).
Table 2. Quantitative MRI Features according to IDH Mutation.
| Imaging Features | IDHmut (n = 47) | IDHwt (n = 29) | P | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| T2 volume | 90.90 | 65.45 | 60.36 | 43.14 | 0.017 |
| CBV_mean | 1.48 | 0.60 | 1.68 | 0.58 | 0.149 |
| CBV_95 | 2.95 | 1.21 | 3.14 | 1.00 | 0.477 |
| CBV_90 | 2.46 | 1.02 | 2.67 | 0.79 | 0.320 |
| CBV_85 | 2.08 | 0.86 | 2.29 | 0.70 | 0.258 |
| CBF_mean | 1.50 | 0.55 | 1.78 | 0.63 | 0.048 |
| CBF_95 | 2.86 | 1.08 | 2.99 | 0.91 | 0.583 |
| CBF_90 | 2.47 | 0.97 | 2.67 | 0.85 | 0.337 |
| CBF_85 | 2.12 | 0.84 | 2.36 | 0.78 | 0.205 |
| ADC_mean | 1.71 | 0.30 | 1.50 | 0.32 | 0.007 |
| ADC_5 | 1.19 | 0.32 | 1.24 | 0.35 | 0.583 |
| ADC_10 | 1.23 | 0.18 | 1.27 | 0.33 | 0.505 |
| ADC_15 | 1.32 | 0.21 | 1.34 | 0.36 | 0.811 |
P values are derived from Student's t and Mann-Whitney U tests.
ADC = apparent diffusion coefficient, CBF = cerebral blood flow, CBV = cerebral blood volume, IDHmut = IDH mutation, IDHwt = IDH wildtype, SD = standard deviation
Table 3. Quantitative MRI Features according to 1p/19q Codeletion in IDH-Mutant Gliomas.
| Imaging Features | 1p/19qdel (n = 19) | 1p/19qnondel (n = 28) | P | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| T2 volume | 76.97 | 59.74 | 100.36 | 68.48 | 0.222 |
| CBV_mean | 1.72 | 0.73 | 1.32 | 0.44 | 0.024 |
| CBV_95 | 3.28 | 1.50 | 2.73 | 0.95 | 0.132 |
| CBV_90 | 2.80 | 1.30 | 2.24 | 0.72 | 0.065 |
| CBV_85 | 2.40 | 1.13 | 1.87 | 0.55 | 0.039 |
| CBF_mean | 1.69 | 0.71 | 1.37 | 0.37 | 0.053 |
| CBF_95 | 3.16 | 1.30 | 2.65 | 0.88 | 0.122 |
| CBF_90 | 2.77 | 1.24 | 2.26 | 0.67 | 0.080 |
| CBF_85 | 2.38 | 1.12 | 1.94 | 0.55 | 0.084 |
| ADC_mean | 1.55 | 0.29 | 1.82 | 0.27 | 0.002 |
| ADC _5 | 1.04 | 0.13 | 1.30 | 0.37 | 0.002 |
| ADC_10 | 1.11 | 0.13 | 1.31 | 0.18 | < 0.001 |
| ADC_15 | 1.19 | 0.15 | 1.42 | 0.20 | < 0.001 |
P values are derived from Student's t and Mann-Whitney U tests. 1p/19del = 1p/19q codeletion, 1p/19qnondel = 1p/19q nondeletion
The univariable logistic regression analysis revealed significant associations between IDH mutation and T2 volume and between IDH mutation and ADC_mean value. Moreover, among the IDHmut tumors, 1p/19q codeletion demonstrated associations with CBV_mean values and ADC_mean, ADC_5, ADC_10, and ADC_15 values (Table 4). The multivariable regression analysis using the results from the univariable regression analysis revealed associations between IDH mutation and ADC_mean as well as 1p/19q codeletion and CBV_mean (Table 4). From the multivariable analysis, the prediction models for the IDH mutation and 1p/19q codeletion using the quantitative imaging parameters were acquired. The diagnostic performance of the prediction model showed accuracy, sensitivity and specificity values of 66.7%, 66.7%, and 72.7%, respectively, for the IDH mutation and of 66.7%, 100.0%, and 50.0%, respectively, for 1p19q codeletion using the validation dataset.
Table 4. Univariable and Multivariable Regression Analyses of Imaging Parameters and Genomic Profiles.
| Univariable Analysis | Multivariable Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| OR | P | OR | P | AUC | 95% CI | P for ROC | Cut-Off | |
| IDH mutation | ||||||||
| T2 volume | 1.01 | 0.035 | 1.01 | 0.139 | ||||
| CBF_mean | 0.45 | 0.058 | ||||||
| ADC_mean | 8.54 | 0.010 | 6.05 | 0.037 | 0.67 | 0.56–0.78 | 0.008 | > 1.49 |
| 1p/19q codeletion | ||||||||
| CBV_mean | 3.49 | 0.037 | 1418.81 | 0.022 | 0.68 | 0.53–0.81 | 0.035 | > 1.59 |
| CBV_85 | 2.22 | 0.059 | ||||||
| ADC_mean | 0.03 | 0.005 | 2.85 | 0.975 | ||||
| ADC_5 | 0.00 | 0.002 | 9.65 | 0.339 | ||||
| ADC_10 | 0.00 | 0.003 | 29.43 | 0.859 | ||||
| ADC_15 | 0.00 | 0.002 | 24.21 | 0.478 | ||||
AUC = area under curve, CI = confidence interval, Cut-off = cut-off values for the prediction of genomic profiles of World Health Organization grade III gliomas, OR = odds ratio, ROC = receiver operating curve
From the comparison of quantitative MRI features between 3 groups of genetic profiles of G3 gliomas, IDHmut1p/19qnondel group revealed the largest tumor volume, lowest CBV_mean, CBV_85, and CBF_mean, as well as the highest ADC_mean, ADC_5, ADC_10, and ADC_15 values (Supplementary Table 4). The IDHmut1p/19qnondel group demonstrated the highest tumor signal intensity ratio between T2WI and FLAIR images, representing the presence of T2-FLAIR mismatch sign from the quantitative analysis (1.46 ± 0.25, 1.53 ± 0.29, and 1.29 ± 0.17 in IDHmut1p/19qdel, IDHmut1p/19qnondel, and IDHwt group G3 gliomas, respectively; p < 0.001).
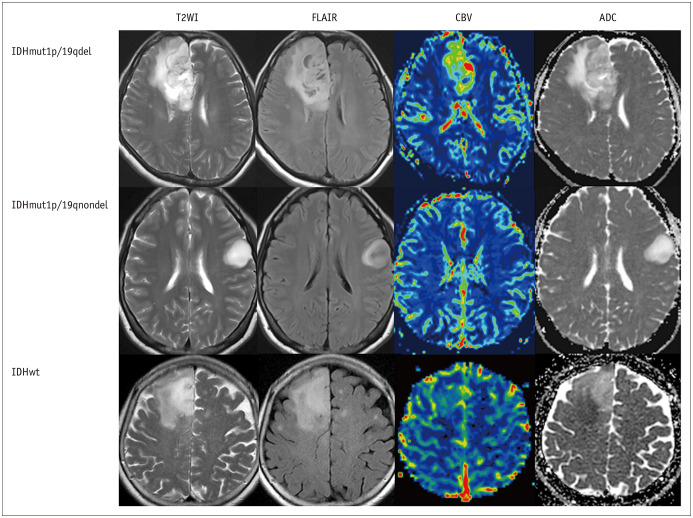
The representative cases of G3 gliomas according to their genomic profiles are depicted in Figure 2.
Fig. 2. Representative cases of G3 gliomas according to their genetic profiles.
A representative case of IDHmut1p/19qdel G3 glioma demonstrated higher CBV and lower ADC values than IDHmut1p/19qnondel tumors. Note that the IDHmut1p/19qnondel group showed a T2-FLAIR mismatch sign, which was defined as a high signal intensity of the tumor in T2WI and relatively low signal intensity on FLAIR, except for the peripheral rim. IDHwt G3 gliomas showed lower ADC values than IDHmut G3 gliomas. ADC = apparent diffusion coefficient, CBV = cerebral blood volume, FLAIR = fluid attenuated inversion recovery, IDH = isocitrate dehydrogenase, IDHmut1p/19qdel = IDH mutation and a chromosome arm 1p/19q-codeleted, IDHmut1p/19qnondel = IDH mutation, 1p/19q-nondeleted, IDHwt = IDH wildtype, T2WI = T2-weighted image
Survival Analysis
The median PFS of the patients included in the study was 38.93 months (95% confidence interval [95CI]: 30.42–43.88 months). The IDHmut1p/19qdel group had the longest median PFS (45.50 months, 95CI: 40.20–50.80). The IDHmut1p/19qnondel group showed a longer median PFS (38.93 months, 95CI: 30.37–49.63) than the IDHwt group (11.90 months, 95CI: 7.70–36.83). The survival curves between the three groups showed statistical significance (p < 0.001) (Fig. 3).
Fig. 3. Kaplan-Meier survival graphs according to genetic profiles.
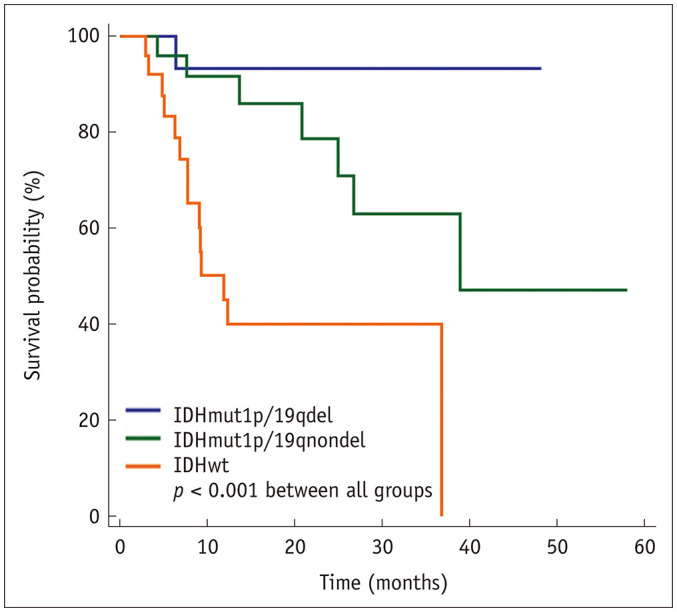
Note that patients with IDHmut1p/19qdel G3 gliomas demonstrate the longest median PFS, followed by patients with IDHmut1p/19qnondel G3 gliomas. Lastly, the IDHwt G3 gliomas had the shortest median PFS. The survival curves between the three groups showed a significant difference (p < 0.001). PFS = progression-free survival
We found statistically significant associations between PFS and the genetic profile of the tumors, such as O6-alkylguanine DNA alkyltransferase (MGMT) promoter methylation status (p = 0.020), CBV_mean (p = 0.010), and CBF_mean (p = 0.029) (Table 5).
Table 5. Multivariable Cox Hazards Regression Analysis of MRI Features with Progression-Free Survival.
| Hazard Ratio | P | |
|---|---|---|
| IDH mutation | 2.25 | 0.495 |
| 1p/19q codeletion | 0.89 | 0.919 |
| ATRX loss | 3.75 | 0.083 |
| MGMT promoter methylation | 0.21 | 0.020 |
| EGFR | 6.67 | 0.185 |
| T2 volume | 0.99 | 0.935 |
| CBV_mean | 0.09 | 0.010 |
| CBV_95 | 0.87 | 0.832 |
| CBV_90 | 1.71 | 0.472 |
| CBV_85 | 1.04 | 0.978 |
| CBF_mean | 9.52 | 0.029 |
| CBF_95 | 2.80 | 0.216 |
| CBF_90 | 0.24 | 0.163 |
| CBF_85 | 0.78 | 0.783 |
| ADC_mean | 2.56 | 0.235 |
| ADC_5 | 25.84 | 0.189 |
| ADC_10 | 7581.34 | 0.566 |
| ADC_15 | 0.08 | 0.111 |
| Enhancement | 5.66 | 0.071 |
| Necrosis | 1.86 | 0.383 |
| Margin | 1.48 | 0.432 |
Comparison between IDHwt GBM and Grade III Gliomas
From the Kaplan-Meier survival analysis of 155 IDHwt GBMs and 29 IDHwt G3 gliomas, the median PFS did not show any significant difference (median PFS: 9.83 vs. 11.90 months, respectively; p = 0.107). From the comparison of quantitative imaging features of IDHwt GBMs and IDHwt G3 gliomas, the CBV_mean (1.74 ± 0.86 vs. 1.75 ± 0.65, p = 0.948) and ADC_mean values (1.48 ± 0.50 vs. 1.36 ± 0.49, p = 0.256) did not reveal any significant differences. However, IDHwt GBMs showed higher tumor volume on T2WI, higher values of CBV_95, CBV_90, and CBV_85, as well as lower ADC_5, ADC_10, and ADC_15 values than IDHwt G3 gliomas (all p values < 0.050).
Interobserver Agreement
Interobserver agreement between the three radiologists revealed an ICC of 0.97 for the CBV_mean and 0.98 for the ADC_mean values, indicating almost perfect agreement between the readers. From Pearson's chi-square test, there was no significant difference in the evaluation of the presence of a T2-FLAIR mismatch sign between the three readers from the interobserver analysis.
DISCUSSION
From this study, we found a significant correlation between qualitative and quantitative MRI features and major genetic and histologic features in G3 gliomas. We found that IDHmut G3 gliomas tend to have more necrosis, a larger tumor volume, lower CBF, and higher ADC values than IDHwt tumors. Among the IDHmut G3 gliomas, the IDHmut1p/19qdel group tended to have more enhancement, higher CBV, and lower ADC values than the IDHmut1p/19qnondel group. The PFS was the longest in the IDHmut1p/19qdel group, followed by the IDHmut1p/19qnondel and IDHwt groups. Additionally, our study discovered that the CBV and CBF values have a significant association with the prognosis of G3 gliomas, regardless of the genetic profiles of the tumors. We found significant associations between the quantitative and qualitative imaging features of G3 gliomas according to the presence of an IDH mutation and 1p/19q codeletion, which were included in the most recent WHO guidelines for glioma classification. With the findings of this study, we were able to create a prediction model. This model can be used in molecular and prognosis prediction of G3 gliomas, providing a better guidance for the patient management of G3 glioma patients.
In LGGs, higher tumor volumes and ADC values for IDHmut tumors than for IDHwt tumors have been reported previously (8,9,14,15). These observations potentially represent the slowly growing nature and lower cellularity of IDHmut G3 gliomas compared to those of IDHwt gliomas. Furthermore, regression analysis revealed significant associations between IDH mutation and ADC values, suggesting the potential role of this particular imaging feature in prediction of IDH mutation status in G3 gliomas. However, the G3 gliomas included in our study did not show a significant difference in the proportions of tumors with contrast-enhancing portions according to the presence of the IDH mutation, inconsistent with findings by Leu et al. (8). This discrepancy might be due to the differences in the number of patients included in the study, since a previous report included only 34 cases of G3 gliomas compared to the 76 cases included in this study. Instead, we found that IDHmut G3 gliomas included in this study showed a high proportion of tumors with necrosis, which is usually present in aggressive tumors. We believe that this observation is due to the increased heterogeneity of IDHmut gliomas as compared to IDHwt (16), which may also result in individual variations from slow growing gliomas with low cellularity to focal aggressive foci within tumors. In addition, previous research has found significantly higher proportions of necrosis in GBMs due to intravascular thrombosis caused by hypoxia-mediated coagulation system activation, resulting in endothelial cell proliferation and necrosis of IDHmut tumors (17,18). Our result are in line with these studies done on GBM, suggesting the similar pathophysiological changes in G3 gliomas.
Among the IDHmut G3 gliomas, a higher proportion of tumors with contrast-enhancing portions, higher CBV and lower ADC values were observed in the IDHmut1p/19qdel group, consistent with a previous report (8) which enhanced the role of these imaging markers in prediction of the genetic profiles of G3 gliomas. Additionally, the regression analysis showed a significant association with the CBV value and the presence of a 1p/19q codeletion in IDHmut G3 gliomas. Since oligodendrogliomas are comprised of only IDHmut1p/19qdel gliomas according to the WHO guidelines (1), our results depicted similar findings to those described in previous studies that reported higher CBV values in oligodendrogliomas compared with astrocytomas (19,20). Also, we found that T2-FLAIR mismatch sign was able to distinguish IDHmut1p/19qnondel G3 gliomas among IDHmut tumors, similar to the result of previous study that included LGGs (11).
From the three group comparison of the MRI features of the G3 gliomas according to their genomic profiles, we have found that IDHmut1p/19qnondel group has a tendency to have higher tumor volume, lower CBV, and higher ADC values compared with IDHmut1p/19qdel and IDHwt groups, similar to that of a previous report (8). However, these findings can only be useful in distinguishing IDHmut1p/19qdel group from IDHmut1p/19qnondel group and IDHmut1p/19qnondel group from IDHwt G3 gliomas. Furthermore, they did not correlate with the prognosis of G3 gliomas. We believe that our stratified approach (prediction of IDH mutation status first, and then to predict 1p/19q codeletion of G3 gliomas) is a more clinically applicable and plausible way to predict the genetic profiles and prognosis of G3 gliomas using MRI features.
In addition, we found significant differences in the prognosis between the three genetic groups of G3 gliomas. Additionally, our data revealed significant associations between prognosis and not only MGMT promoter methylation status, but also imaging parameters, such as CBV and CBF, which are consistent with previous reports (21,22). This enhances support for the function of imaging parameters as promising biomarkers for predicting the prognosis of G3 glioma patients. Interestingly, the direction of CBV and CBF in relation to survival was opposite from the results of our study, which was similar to one previous study that have analyzed associations between survival and perfusion/diffusion parameters. Also, this previous report did not find a significant association between ADC and survival in LGGs, which were consistent with the results of this study (22). Unexpectedly, IDH mutation and 1p/19q codeletion status were not significantly associated with survival according to multivariable analysis. We believe that further studies with a large data set is needed to demonstrate the role of multiple variables such as genetic profiles and imaging features in predicting survival in G3 glioma patients.
Furthermore, our data depicted that there was no significant difference in the prognosis of IDHwt G3 gliomas and GBMs, which is in concordance with previous reports (7,23,24). Additionally, the imaging parameters of IDHwt G3 gliomas and GBMs were similar to one another, with a wider distribution of values in GBMs due to the heterogeneity of these tumors. Recently, a new classification of LGG, known as the Consortium to Inform Molecular and Practical Approaches to central nervous system Tumor Taxonomy (cIMPACT-NOW), has been introduced and states that only a subset of LGG with specific molecular subtype can be termed as IDHwt, with molecular features of GBM, that follow the prognosis of IDHwt GBMs (25). Therefore, we believe that there is a need for a future study that includes the specific subset of IDHwt LGGs according to the new classification stated by cIMPACT-NOW, which investigates their imaging features compared with IDHwt GBMs.
There are some limitations in this study beyond the nature of its retrospective design. First, the patients in this study underwent different durations of radiation therapy, which might have affected the PFS of the G3 glioma patients included in the study. Second, we analyzed MR images from different MR scanners from various manufacturers with different field strengths. However, we normalized the CBV, CBF, and ADC values to the normal white matter area of the corona radiata on the contralateral side of the tumor to diminish the potential bias of the CBV and ADC measurements. Third, some genetic information was missing for a small number of patients. However, the number of patients with missing genetic information was small. Fourth, we included internal cystic and necrotic lesions of the tumor for analysis to identify features and to develop a model that will represent the entire tumor characteristics rather than parts of the tumor. In effect, our results might contradict those of previous studies. However, we believe that since the majority of the tumors included in the study contained relatively large portions of cystic or necrotic areas, segmenting all parts of the tumor was more feasible for the analysis and future application of the prediction model. Fifth, we only included G3 gliomas for analysis in this study, which is slightly different from studies that included LGGs (G2 and G3 gliomas). So far, our study is the only study with enough number of G3 glioma patients included that evaluated quantitative characteristics of G3 gliomas, and we believe this gives a unique value to this study.
In conclusion, we found significant differences in the MRI features in G3 gliomas according to different molecular subtypes, which can be utilized for the prediction of the genomic profiles and prognosis of G3 glioma patients. Additionally, the MRI signatures and prognosis of IDHwt G3 gliomas tend to follow those of IDHwt GBMs. We believe that our application can be used in precision medicine for the diagnosis and treatment planning for G3 glioma patients.
Footnotes
This study was supported by a grant from the Korea Healthcare technology R&D Projects, Ministry for Health, Welfare & Family Affairs (HI16C1111), by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2016M3C7A1914002), by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2020R1A2C2008949 and NRF-2020R1A4A1018714), by Creative-Pioneering Researchers Program through Seoul National University (SNU), and by the Institute for Basic Science (IBS-R006-A1).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Supplementary Materials
The Data Supplement is available with this article at https://doi.org/10.3348/kjr.2020.0011.
MR Scan Parameters
Imaging Parameter Descriptions
Qualitative MRI Features according to Histologic Classification and Genetic Profiles
Quantitative MRI Features according to Genetic Profiles
References
- 1.Hoshide R, Jandial R. 2016 World Health Organization classification of central nervous system tumors: an era of molecular biology. World Neurosurg. 2016;94:561–562. doi: 10.1016/j.wneu.2016.07.082. [DOI] [PubMed] [Google Scholar]
- 2.van den Bent MJ. Practice changing mature results of RTOG study 9802: another positive PCV trial makes adjuvant chemotherapy part of standard of care in low-grade glioma. Neuro Oncol. 2014;16:1570–1574. doi: 10.1093/neuonc/nou297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ichimura K, Pearson DM, Kocialkowski S, Bäcklund LM, Chan R, Jones DT, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 6.Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, et al. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Brat DJ, Verhaak RGW, Aldape KD, Yung WKA, Salama SR, Cooper LAD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leu K, Ott GA, Lai A, Nghiemphu PL, Pope WB, Yong WH, et al. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II–III diffuse gliomas. J Neurooncol. 2017;134:177–188. doi: 10.1007/s11060-017-2506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CC, Jain R, Radmanesh A, Poisson LM, Guo WY, Zagzag D, et al. Predicting genotype and survival in glioma using standard clinical MR imaging apparent diffusion coefficient images: a pilot study from the cancer genome atlas. AJNR Am J Neuroradiol. 2018;39:1814–1820. doi: 10.3174/ajnr.A5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batchala PP, Muttikkal TJE, Donahue JH, Patrie JT, Schiff D, Fadul CE, et al. Neuroimaging-based classification algorithm for predicting 1p/19q-codeletion status in IDH-mutant lower grade gliomas. AJNR Am J Neuroradiol. 2019;40:426–432. doi: 10.3174/ajnr.A5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M, Han K, Ahn SS, Bae S, Choi YS, Hong JB, et al. The added prognostic value of radiological phenotype combined with clinical features and molecular subtype in anaplastic gliomas. J Neurooncol. 2019;142:129–138. doi: 10.1007/s11060-018-03072-0. [DOI] [PubMed] [Google Scholar]
- 13.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 14.Xing Z, Yang X, She D, Lin Y, Zhang Y, Cao D. Noninvasive assessment of IDH mutational status in World Health Organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. AJNR Am J Neuroradiol. 2017;38:1138–1144. doi: 10.3174/ajnr.A5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metellus P, Coulibaly B, Colin C, de Paula AM, Vasiljevic A, Taieb D, et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010;120:719–729. doi: 10.1007/s00401-010-0777-8. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Choi SH, Ryoo I, Yoon TJ, Kim TM, Lee SH, et al. Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neurooncol. 2015;121:141–150. doi: 10.1007/s11060-014-1614-z. [DOI] [PubMed] [Google Scholar]
- 17.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 18.Homma T, Fukushima T, Vaccarella S, Yonekawa Y, Di Patre PL, Franceschi S, et al. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol. 2006;65:846–854. doi: 10.1097/01.jnen.0000235118.75182.94. [DOI] [PubMed] [Google Scholar]
- 19.Cha S, Tihan T, Crawford F, Fischbein NJ, Chang S, Bollen A, et al. Differentiation of low-grade oligodendrogliomas from low-grade astrocytomas by using quantitative blood-volume measurements derived from dynamic susceptibility contrast-enhanced MR imaging. AJNR Am J Neuroradiol. 2005;26:266–273. [PMC free article] [PubMed] [Google Scholar]
- 20.Lev MH, Ozsunar Y, Henson JW, Rasheed AA, Barest GD, Harsh GR, 4th, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected] AJNR Am J Neuroradiol. 2004;25:214–221. [PMC free article] [PubMed] [Google Scholar]
- 21.Law M, Young RJ, Babb JS, Peccerelli N, Chheang S, Gruber ML, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology. 2008;247:490–498. doi: 10.1148/radiol.2472070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latysheva A, Emblem KE, Server A, Brandal P, Meling TR, Pahnke J, et al. Survival associations using perfusion and diffusion magnetic resonance imaging in patients with histologic and genetic defined diffuse glioma World Health Organization grades II and III. J Comput Assist Tomogr. 2018;42:807–815. doi: 10.1097/RCT.0000000000000742. [DOI] [PubMed] [Google Scholar]
- 23.Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129:585–596. doi: 10.1007/s00401-015-1398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuss DE, Mamatjan Y, Schrimpf D, Capper D, Hovestadt V, Kratz A, et al. IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol. 2015;129:867–873. doi: 10.1007/s00401-015-1438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis DN, Ellison DW, Brat DJ, Aldape K, Capper D, Hawkins C, et al. cIMPACT-NOW: a practical summary of diagnostic points from Round 1 updates. Brain Pathol. 2019;29:469–472. doi: 10.1111/bpa.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MR Scan Parameters
Imaging Parameter Descriptions
Qualitative MRI Features according to Histologic Classification and Genetic Profiles
Quantitative MRI Features according to Genetic Profiles