Fig. 3. UPRmt requires the weak MTS in ATFS-1.
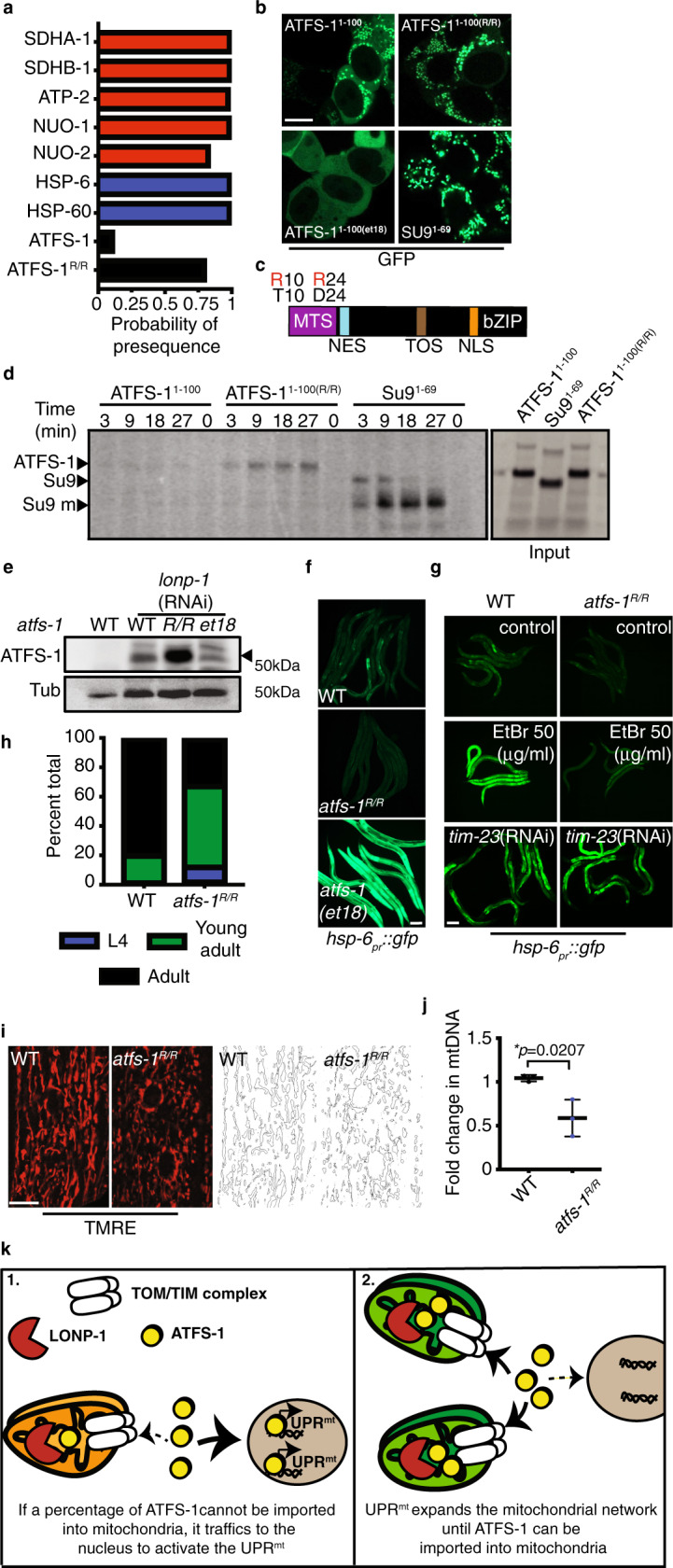
a Mitochondrial targeting sequence probability prediction using MitoFates. OXPHOS proteins (red), mitochondrial chaperones (blue), ATFS-1 (black). b HEK293T cells expressing ATFS-11–100::GFP, ATFS-11-100(R/R)::GFP, ATFS-11-100(et18)::GFP or SU91–69::GFP. Scale bar 10 µm. Experiment was repeated two biologically independent times with similar results. c ATFS-1 schematic highlighting ATFS-1R/R amino acid substitutions. d In organelle import of radiolabeled ATFS-11–100::GFP, ATFS-11-100(R/R)::GFP and Su91–69::GFP into isolated mitochondria. After the indicated time points, mitochondria were washed and analyzed by SDS-PAGE electrophoresis. ATFS-1, Su9, mature (m) are marked. N = 3 biologically independent experiments with similar results. e SDS-Page immunoblots of lysates from wild-type, atfs-1R/R, and atfs-1(et18) worms raised on control or lonp-1(RNAi). ATFS-1 is marked (‣) and tubulin (Tub) was used as a loading control. N = 3 biologically independent experiments with similar results. f hsp-6pr::gfp in wild-type, atfs-1(et18), and atfs-1R/R worms. Scale bar 0.1 mm. N = 3 biologically independent experiments with similar results. g hsp-6pr::gfp in wild-type and atfs-1R/R worms raised on 50 µg/ml EtBr, control, or timm-23(RNAi). Scale bar 0.1 mm. N = 3 biologically independent experiments with similar results. h Developmental stages of 3-day-old wild-type or atfs-1R/R worms. N = 158 worms (wild type) and N = 256 worms (atfs-1R/R). Experiments were repeated two biologically independent with similar results. i TMRE staining of wild-type and atfs-1R/R worms. Skeleton-like binary backbone is presented (right). Scale bar 10 µm. Experiments were repeated three times with similar results. j Quantification of mtDNA in wild-type and atfs-1R/R worms as determined by qPCR. N = 3 biologically independent experiments. Error bars mean ± SD (two-tailed Student’s t-test). k Proposed model for ATFS-1-mediated mitochondria expansion. Source data are provided as a Source Data file.
