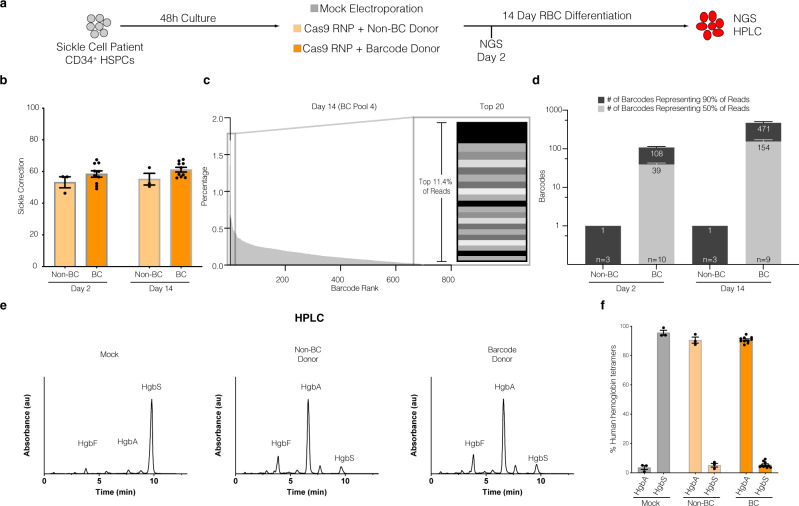
Fig. 2. Correction of the Sickle Cell Disease-causing E6V mutation using barcoded AAV6 donors in SCD-derived CD34+ HSPCs.
a Experimental design – SCD patient-derived CD34+ HSPCs edited with CRISPR/Cas9 RNP and electroporation only (mock), single donor (non-BC), or barcode donor (BC) AAV6 HR templates. b SCD correction efficiency (percentage of corrected sickle cell alleles) of non-BC and BC treated groups as a fraction of total NGS reads (e.g., HR reads / [all reads].) n = 3 (non-BC) and n = 10 (BC) biologically independent replicates examined over 3 independent experiments. c Representative example of barcode fractions in descending order from one donor at day 14 time point. Right: Top 20 clones represented as stacked bar graph (representing 11.4% of reads). d Number of unique barcode alleles comprising the top 50% and top 90% of reads from each treatment condition, sampling approximately 1000 cells per condition (see Supplementary Tables 1 and 2). e Representative hemoglobin tetramer HPLC chromatograms of RBC differentiated cell lysates at day 14 post-treatment. f Quantification of total hemoglobin protein expression in each group. Each data point represents an individual biological replicate. n = 3 (Mock and non-BC) and n = 10 (BC) biologically independent replicates examined over 2 independent experiments. HgbA adult hemoglobin, HgF fetal hemoglobin, HbS sickle hemoglobin, AAV6 Recombinant AAV2/6 vector. Error bars depict mean ± SEM.