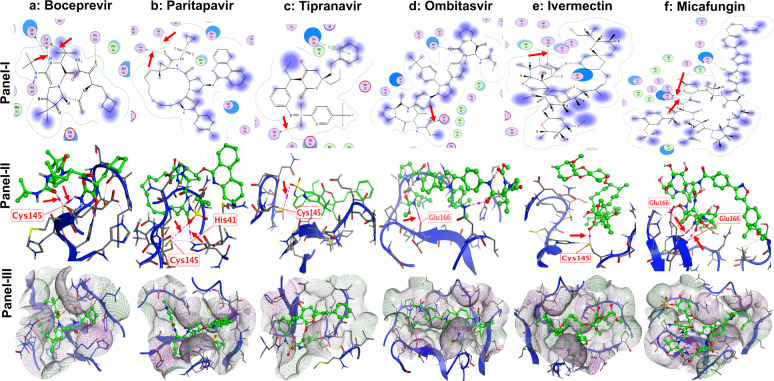
Fig. 6. Structural analysis and interaction of the potential inhibitors with SARS-CoV-2 3CLpro enzyme.
Inhibitors were docked with active site of 3CLpro protein. Panel-I: ligand interaction map. Panel-II: interaction of inhibitors specific amino acids of 3CLpro at active site. Panel-III: lipophilic cavity of active site with drugs interacting with specific amino acids. Boceprevir (a), partapravir (b), tipranavir (c), ombitasvir (d), ivermectin (e), and micafungin (f) are arranged in columns for comparison. Drugs are represented in green color with ball and stick model. Arrows indicate the C–H, N–H, and C–O bonds between drugs and with Cys145, His41, and Glu166 residues since they are essential for the enzymatic activity of 3CLpro enzyme. However, we also observed the drugs interacting with neighboring amino acid residues.