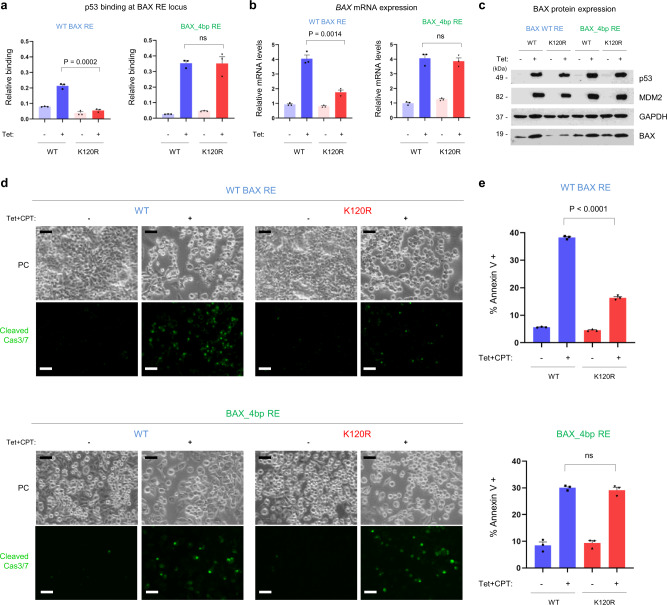
Fig. 5. Genomic editing of BAX RE modifies p53 binding and interconverts cell fate outcome.
a ChIP signals at the genomic BAX RE locus for WT (blue) and K120R (red) p53 in cells with a WT BAX RE (left) or genetically edited BAX_4bp RE (right). Graphs represent precipitated DNA relative to an input (total) DNA and show cumulative data from three independent experiments (mean ± s.e.m.). b Quantitative polymerase chain reaction (qPCR) of BAX mRNA (represented relative to GAPDH mRNA) from the same cells as in panel a. The graphs show cumulative data from three independent experiments (mean ± s.e.m.). c Representative Western blot from three independent experiments. Blots were probed with antibodies specific for p53, MDM2, GAPDH, and BAX proteins. d Same cells as in panels a–c were treated additionally for 12 h with camptothecin (CPT, 1 μM; to induce DNA damage) and induction of apoptosis was detected by visualizing cleaved Caspase-3/7 by fluorescence microscopy (d), or by measuring Annexin V positivity by flow cytometry (e). In both assays, a restoration of pro-apoptotic functions of the K120R-p53 variant in cells with edited BAX RE could be observed. Scale bars, 50 μm. PC, phase contrast. Representative images from three independent experiments are shown in panel d. Graphs in panel e represent an average percentage of Annexin V-positive cells from three independent experiments (mean ± s.e.m.). Two-tailed unpaired Students t-test was used for statistical analysis in panels a, b, and e. Source data are provided as a Source Data file.