Abstract
Heterobasidion species are amongst the most intensively studied polypores because several species are aggressive white rot pathogens of managed coniferous forests mainly in Europe and North America. In the present study, both morphological and multilocus phylogenetic analyses were carried out on Heterobasidion samples from Asia, Oceania, Europe and North America. Three new taxa were found, i.e., H. armandii, H. subinsulare, and H. subparviporum are from Asia and are described as new species. H. ecrustosum is treated as a synonym of H. insulare. So far, six taxa in the H. annosum species complex are recognized. Heterobasidion abietinum, H. annosum, and H. parviporum occur in Europe, H. irregulare, and H. occidentale in North America, and H. subparviporum in East Asia. The North American H. irregulare was introduced to Italy during the Second World War. Species in the H. annosum complex are pathogens of coniferous trees, except H. subparviporum that seems to be a saprotroph. Ten species are found in the H. insulare species complex, all of them are saprotrophs. The pathogenic species are distributed in Europe and North America; the Asian countries should consider the European and North American species as entry plant quarantine fungi. Parallelly, European countries should consider the American H. occidentale and H. irregulare as entry plant quarantine fungi although the latter species is already in Italy, while North America should treat H. abietinum, H. annosum s.s., and H. parviporum as entry plant quarantine fungi. Eight Heterobasidion species found in the Himalayas suggest that the ancestral Heterobasidion species may have occurred in Asia.
Keywords: taxonomy, phylogeny, new taxa, Bondarzewiaceae, pathogenic fungi
Introduction
The polypore genus Heterobasidion Bref., which belongs to the family Bondarzewiaceae, is one of the most intensively studied basidiomycetous genera because some species of Heterobasidion are aggressive pathogens of managed coniferous forests in Europe and North America (Woodward et al., 1998). Two morphological taxa, H. annosum (Fr.) Bref. and H. insulare (Murrill) Ryvarden, had generally been accepted in Heterobasidion (Murrill, 1908; Gilbertson and Ryvarden, 1986; Ryvarden and Gilbertson, 1993; Núñez and Ryvarden, 2001). However, mating studies have revealed that both H. annosum and H. insulare are in fact species complexes (Korhonen, 1978; Dai and Korhonen, 1999; Dai et al., 2002, 2003).
Three species, Heterobasidion abietinum Niemelä and Korhonen (Eur F-group), H. annosum (Fr.) Bref. sensu stricto (Eur P-group) and H. parviporum Niemelä and Korhonen (Eur S-group), have been recognized in Europe (Niemelä and Korhonen, 1998), and two species, H. irregulare Garbel. and Otrosina (NAm P-group) and H. occidentale Otrosina and Garbel. (NAm S-group), were described from North America (Otrosina and Garbelotto, 2010). Based on mating studies, the East Asian taxon in the H. annosum species complex was considered as H. parviporum (Dai and Korhonen, 1999, 2003; Dai et al., 2006; Dai, 2012; Chen et al., 2015). Similarly, investigations based on mating tests, morphological characteristics and molecular analyses revealed several species also within the Asian H. insulare complex: H. linzhiense Y. C. Dai and Korhonen (Dai et al., 2007), H. australe Y. C. Dai and Korhonen (2009), H. ecrustosum Tokuda, T. Hatt. and Y. C. Dai, H. orientale Tokuda, T. Hatt. and Y. C. Dai (Tokuda et al., 2009), H. amyloideum Y. C. Dai, Jia J. Chen and Korhonen, H. tibeticum Y. C. Dai, Jia J. Chen and Korhonen (Chen et al., 2014) and H. amyloideopsis Saba, C. L. Zhao, Khalid and Pfister (Zhao et al., 2017). In addition, H. araucariae P. K. Buchanan from Australia and adjacent regions (Buchanan, 1988) was confirmed to be a member of the H. insulare species complex (Chen et al., 2015).
Earlier phylogenetic analyses on the H. annosum complex used sequences of the internal transcribed spacer (ITS) and intergenic spacer (IGS) regions of the nuclear genes, and manganese peroxidase genes, and laccase genes (Maijala et al., 2003; Asiegbu et al., 2004). Later, several attempts were made to resolve the taxonomy of the H. annosum complex or H. insulare complex using multilocus phylogenetic approaches (Johannesson and Stenlid, 2003; Ota et al., 2006; Linzer et al., 2008; Chen et al., 2014). Recently, five species in the H. annosum species complex and eight species in H. insulare species complex were also recognized and confirmed by multilocus phylogenetic approaches, and divided into three groups based on five nuclear genes and two mitochondrial genes, i.e., ITS, the large nuclear ribosomal RNA subunit (nrLSU), the largest subunit of RNA polymerase II (RPB1), the second subunit of RNA polymerase II (RPB2), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), mitochondrial ATP synthase subunit 6 (ATP6), and mitochondrial small subunit rDNA (mtSSU) (Chen et al., 2015).
Several hypotheses on the evolutionary scenarios of the Heterobasidion have been put forward (Otrosina et al., 1993; Ota et al., 2006; Linzer et al., 2008). Dalman et al. (2010) proposed that the H. annosum complex originated in Laurasia, H. annosum s.s./H. irregulare arose in Eurasia, and H. parviporum/H. abietinum/H. occidentale, which occurred in eastern Asia or western North America, emerged between 45 and 60 Ma in the Palaearctic; this conclusion was based on non-coding regions of elongation factor 1-α (EFA), glutathione-S-transferase (GST1), GAPDH, and transcription factor (TF). Recently, based on more species and samples of Heterobasidion and the fossil record, molecular dating suggested that ancestral Heterobasidion species originated in Eurasia occurred mainly during the Early Miocene (Chen et al., 2015; Zhao et al., 2017).
Based on a larger set of Heterobasidion samples from Asia, Oceania, Europe and North America, and using combined RPB1 and RPB2 sequence dataset, a further phylogenetic investigation on the genus is carried out. Four new taxa are detected, and three of them are described and illustrated in the present paper. Moreover, most relevant morphological characteristics of different species of Heterobasidion are compared.
Materials and Methods
Morphological Studies
The studied specimens and cultures (Table 1) are deposited at the herbaria of Institute of Microbiology of the Beijing Forestry University (BJFC, Beijing, China), Natural Resources Institute Finland (Luke, Helsinki, Finland), U.S. Forest Service, Northern Research Station (CFMR, Madison, WI, United States), private herbarium of J. Vlasák (JV, České Budějovice, Czechia), and Landcare Research, New Zealand (PDD, Lincoln, New Zealand). Ecology and some macromorphological characters were based on field notes. Anatomy was studied, and measurements and drawings were made from slide preparations stained with Cotton Blue. Drawings were made with the aid of a drawing tube. In presenting the variation in the size of the spores, the 5% of the measurements at each end of the range are shown in parentheses. Basidiospore spine lengths are not included in the measurements. The following abbreviations are used: IKI = Melzer’s reagent, IKI− = both non-amyloid and non-dextrinoid, IKI+ = amyloid, KOH = 5% potassium hydroxide, CB = Cotton Blue, CB+ = cyanophilous, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios between the specimens studied, n = number of spores measured from given number of specimens. Color terms are from Petersen (1996).
TABLE 1.
Information on samples of Heterobasidion used in this study.
| Species | Sample no. | Geographic origin | Host | GenBank accessions |
||||
| ITS | nrLSU | RPB1 | RPB2 | GAPDH | ||||
| Bondarzewia occidentalis | HHB 14803 | Washington, United States | Picea sitchensis | DQ200923 | DQ234539 | DQ256049 | AY218474 | – |
| B. submesenterica | Cui 10724 | Sichuan, China | Abies ernestii | KJ583205 | KJ583219 | KJ651627 | KJ651720 | KJ651752 |
| Heterobasidion abietinum | 00053/1 | Trentino, Italy | Picea abies | KJ651451 | KJ651509 | KJ651630 | KJ651723 | KJ651754 |
| H. abietinum | 00055/6 | Trentino, Italy | Picea abies | KJ651452 | KJ651510 | KJ651631 | KJ651724 | KJ651755 |
| H. abietinum | 00057/2 | Trentino, Italy | Abies alba | KJ651453 | KJ651511 | KJ651632 | KJ651725 | KJ651756 |
| H. amyloideum | Cui 12656 | Tibet, China | Pinus | MT146480 | MT446029 | MT157738 | MT157761 | MT157721 |
| H. amyloideum | Cui 12274 | Tibet, China | Abies | MT146481 | MT446030 | MT157739 | MT157762 | MT157722 |
| H. annosum | 06071/1 | Lazio, Italy | Pinus pinea | KJ651458 | KJ651516 | KF453497 | KF453491 | KJ651761 |
| H. annosum | 06125/2 | Krasnoyarsk, Russia | Pinus sylvestris | KJ651459 | KJ651517 | KF453498 | KF453492 | KJ651762 |
| H. annosum | 06129/6 | Krasnoyarsk, Russia | Pinus sylvestris | KJ583211 | KJ583225 | KF006499 | KF033133 | KJ651763 |
| H. araucariae | 65008 | Queensland, Australia | Araucaria cunninghamii | KJ651462 | KJ651520 | KJ651636 | KJ651729 | KJ651766 |
| H. araucariae | 82001 | Queensland, Australia | Araucaria cunninghamii | KJ651463 | KJ651521 | KJ651637 | KJ651730 | KJ651767 |
| H. armandii | Dai 17605 | Yunnan, China | Pinus armandii | MT146482 | MT446031 | MT157740 | MT157763 | – |
| H. armandii | Dai 17606 | Yunnan, China | Pinus armandii | MT146483 | MT446032 | MT157741 | MT157764 | – |
| H. armandii | Dai 17607 | Yunnan, China | Pinus armandii | MT146484 | MT446033 | MT157742 | MT157765 | – |
| H. australe | Cui 12602 | Yunnan, China | Pinus sp. | MT146485 | MT446034 | MT157743 | MT157766 | MT157723 |
| H. australe | Dai 13507 | Yunnan, China | Pinus sp. | MT146486 | MT446035 | MT157744 | MT157767 | MT157724 |
| H. australe | Dai 13863 | Yunnan, China | Pinus sp. | MT146487 | MT446036 | MT157745 | MT157768 | MT157725 |
| H. insulare | FPRI 429 | Philippines | Pinus sp. | MT146488 | MT446037 | MT157746 | MT157769 | MT157726 |
| H. insulare | Dai 13933 | Chongqing, China | Pinus massoniana | MT146489 | MT446038 | MT157747 | MT157770 | MT157727 |
| H. insulare | Dai 15095 | Jiangxi, China | Pinus massoniana | MT146490 | MT446039 | MT157748 | MT157771 | MT157728 |
| H. irregulare | 57001/TI | North Carolina, United States | Pinus strobus | KJ651473 | KJ651531 | KJ651638 | KJ651731 | KJ651777 |
| H. irregulare | 88010/1 | Vermont, United States | Pinus sp. | KJ651475 | KJ651533 | KJ651640 | KJ651733 | KJ651779 |
| H. irregulare | 01062 | Ontario, Canada | Pinus resinosa | KJ651477 | KJ651535 | KJ651642 | KJ651735 | KJ651781 |
| H. linzhiense | Cui 7216 | Sichuan, China | Abies sp. | KJ651480 | KJ651538 | KF006524 | KF033148 | KJ651784 |
| H. linzhiense | Cui 9645 | Tibet, China | Picea sp. | KJ651481 | KJ651539 | KF033147 | KF006523 | KJ651785 |
| H. linzhiense | Dai 5408 | Tibet, China | Picea sp. | KJ651484 | KJ651542 | KF033154 | KF006533 | KJ651788 |
| H. occidentale | 79034/TI | Alaska, United States | Picea sp. | KJ651485 | KJ651543 | KJ651645 | KJ651738 | KJ651789 |
| H. occidentale | 98004/TI | Oregon, United States | Picea engelmannii | KJ651488 | KJ651546 | KJ651648 | KJ651741 | KJ651792 |
| H. occidentale | 98005/TI | Oregon, United States | Abies magnifica var. shastensis | KJ651489 | KJ651547 | KJ651649 | KJ651742 | KJ651793 |
| H. orientale | Cui 11637 | Heilongjiang, China | Unknown | MT146491 | MT446040 | MT157749 | MT157772 | MT157729 |
| H. orientale | Cui 11815 | Heilongjiang, China | Pinus sp. | MT146492 | MT446041 | MT157750 | MT157773 | MT157730 |
| H. orientale | Cui 12026 | Heilongjiang, China | Picea sp. | MT146493 | MT446042 | MT157751 | MT157774 | MT157731 |
| H. parviporum | 04121/3 | Artjärvi, Finland | Picea abies | KJ583212 | KJ583226 | KF453493 | KF453499 | KJ651800 |
| H. parviporum | 08021/7 | Krasnoyarsk, Russia | Picea abies | KJ651498 | KJ651556 | KF453494 | KF453500 | KJ651803 |
| H. parviporum | 08123/TI | Irkutsk, Russia | Picea abies | KJ651500 | KJ651558 | KF453495 | KF453501 | KJ651805 |
| H. sp. | Korhonen 05030 | California, United States | Pinus ponderosa | MT146494 | MT446043 | MT157752 | MT157775 | – |
| H. sp. | Korhonen 05038 | California, United States | Pinus ponderosa | MT146495 | MT446044 | MT157753 | MT157776 | – |
| H. sp. | Korhonen 05039 | California, United States | Pinus ponderosa | MT146496 | MT446045 | MT157754 | MT157777 | – |
| H. subinsulare | Dai 13842 | Yunnan, China | Pinus sp. | MT146497 | MT446046 | MT157755 | MT157778 | MT157732 |
| H. subinsulare | Li 140804-30 | Yunnan, China | Pinus sp. | MT146498 | MT446047 | MT157756 | MT157779 | MT157733 |
| H. subparviporum | Cui 6961 | Hubei, China | Abies fargesii | KJ651504 | KJ651562 | KJ651658 | KJ651751 | KJ651809 |
| H. subparviporum | Cui 9267 | Tibet, China | Picea sp. | MT146499 | MT446048 | MT157757 | MT157780 | MT157734 |
| H. subparviporum | Dai 14803 | Jilin, China | Picea sp. | MT146500 | MT446049 | MT157758 | MT157781 | MT157735 |
| H. tibeticum | Cui 12257 | Tibet, China | Pinus sp. | MT146501 | MT446050 | MT157759 | MT157782 | MT157736 |
| H. tibeticum | Cui 12335 | Tibet, China | Pinus sp. | MT146502 | MT446051 | MT157760 | MT157783 | MT157737 |
New sequences are shown in bold.
DNA Extraction, PCR Amplification and Sequencing
The Rapid Plant Genome kit based on acetyl trimethylammonium bromide extraction (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to extract genomic DNA from dried fungal specimens and cultures, according to the manufacturer’s instructions with some modifications (Chen et al., 2014). The PCR primers for all genes are listed in Table 2. The PCR procedure for nrLSU was as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 30 s, 50°C for 1 min, 72°C for 1.5 min, and a final extension at 72°C for 10 min. The following PCR protocol for GAPDH, and ITS was used: initial denaturation at 95°C for 3 min, followed by 35 cycles at 94°C for 40 s, (50°C for GAPDH, 54°C for ITS), 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR procedure for RPB1 and RPB2 followed Justo and Hibbett (2011) with slight modifications: initial denaturation at 94°C for 2 min, followed by 10 cycles at 94°C for 40 s, 60°C for 40 s, 72°C for 2 min, then followed by 37 cycles at 94°C for 45 s, 55°C for 1.5 min and 72°C for 2 min, and a final extension at 72°C for 10 min. PCR products were purified with a Gel Extraction and PCR Purification Combo Kit (Spin-column) in Beijing Genomics Institute, Beijing, China. The purified products were then sequenced on an ABI-3730-XL DNA Analyzer (Applied Biosystems, Foster City, CA, United States) using the same primers as in the original PCR amplifications. All newly generated sequences were deposited at GenBank1 and listed in Table 1.
TABLE 2.
PCR primers used in this study.
| Gene | Primer | Primer sequences (5′-3′)a | References |
| GAPDH | GAPDH-F | YGG TGT CTT CAC CAC CAC YGA SSA | Johannesson et al., 2000 |
| GAPDH-R | RTA NCC CCA YTC RTT RTC RTA CCA | Johannesson et al., 2000 | |
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | White et al., 1990 |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | White et al., 1990 | |
| nrLSU | LROR | ACC CGC TGA ACT TAA GC | Vilgalys and Hester, 1990 |
| LR7 | TAC TAC CAC CAA GAT CT | Vilgalys and Hester, 1990 | |
| RPB1 | RPB1-Af | GAR TGY CCD GGD CAY TTY GG | Matheny et al., 2002 |
| RPB1-Cf | CCN GCD ATN TCR TTR TCC ATR TA | Matheny et al., 2002 | |
| RPB2 | fRPB2-5F | GAY GAY MGW GAT CAY TTY GG | Liu et al., 1999; Matheny, 2005 |
| fRPB2-7cR | CCC ATR GCT TGY TTR CCC AT | Liu et al., 1999; Matheny, 2005 |
aDegeneracr codes: S = G or C, W = A or T, R = A or G, Y = C or T, N = A or T or C or G, D = G or A or T, M = A or C.
Phylogenetic Analysis
Bondarzewia occidentalis Jia J. Chen, B. K. Cui and Y. C. Dai and B. submesenterica Jia J. Chen, B. K. Cui and Y. C. Dai were used as outgroups (Chen et al., 2015). Sequences were aligned with BioEdit (Hall, 1999) and ClustalX (Thompson et al., 1997). Sequence alignments were deposited at TreeBase2 (submission ID 25908).
Maximum parsimony (MP) analysis was applied to single-locus genealogies for ITS, nrLSU, RPB1, PPB2, and GAPDH, and combination datasets that contained the RPB1-RPB2 sequences. The tree construction procedure was performed in PAUP∗ version 4.0b10 (Swofford, 2002). All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed, and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap analysis with 1000 replicates (Felsenstein, 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RCI), and homoplasy index (HI), were calculated for each maximum parsimonious tree generated. Phylogenetic trees were visualized using Treeview (Page, 1996).
MrMODELTEST2.3 (Nylander, 2004) was used to determine the best-fit evolution model for the combined dataset for Bayesian inference (BI). The BI was calculated with MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) with a general time reversible model of DNA substitution and an invgamma distribution rate variation across sites. Eight Markov chains were run from random starting tree for 1 M generations of RPB1 and RPB2 dataset, and sampled every 100 generations. The burn-in was set to discard the first 25% of the trees. A majority rule consensus tree of all remaining trees was calculated. Branches that received bootstrap values for MP and Bayesian posterior probabilities (BPP) greater than or equal to 75% (MP) and 0.95 (BPP) were considered as significantly supported.
To determine if the datasets were significantly conflicted, the partition homogeneity test option in PAUP 4.0b was used between the loci in all possible pairwise combinations using 1000 replicates and the heuristic general search option. This test randomly shuffles phylogenetically informative sites between two paired loci: if the datasets are compatible, shuffling sites between the loci should not produce summed tree lengths that are significantly greater than those produced by the observed data (Farris et al., 1994; Huelsenbeck et al., 1996).
Results
Molecular Phylogeny
All targeted DNA loci were successfully amplified and sequenced from our Heterobasidion samples and the outgroup species. Partition homogeneity test showed no conflicts for the RPB1 and RPB2 combined loci (P = 0.019, P ≥ 0.01). Therefore, the amino acid sequences from RPB1 and RPB2 were combined into a single sequence set. The combined dataset included sequences from 46 specimens representing 18 species. The dataset had an aligned length of 2505 characters, of which 1796 characters were constant, 671 were variable and parsimony-uninformative, and 38 were parsimony-informative. The maximum parsimony analysis yielded four equally parsimonious tree (TL = 1033, CI = 0.789, HI = 0.925, RI = 0.730, RC = 0.211). The best model for the combined RPB1 + RPB2 estimated and applied in the Bayesian analysis: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). The Bayesian analysis resulted in a topology similar to the MP analysis, with an average standard deviation of split frequencies = 0.006737, and only the MP tree was provided. Both bootstrap values (>50%) and BPPs (≥0.90) were shown at the nodes (Figure 1).
FIGURE 1.
Phylogeny of Heterobasidion and related species generated by Maximum Parsimony based on combined RPB1 + RPB2 sequences. Parsimony bootstrap values (before the slash markers) higher than 50% and Bayesian posterior probabilities (after the slash markers) more than 0.95 were indicated along branches.
Three new species, Heterobasidion armandii, H. subinsulare, and H. subparviporum formed a well-supported phylogenetic lineages, respectively (100% MP, and 1 BPPs), and phylogenetically distinct from other known species of Heterobasidion.
According to the present phylogenetic analyses, Heterobasidion spp. consists of three lineages: (1) the lineage associated to pines, firs and spruces (H. amyloideopsis, H. amyloideum, H. araucariae, H. armandii, H. australe, H. insulare, H. linzhiense, H. orientale, H. subinsulare, and H. tibeticum); (2) lineage mainly associated to pines (H. annosum s.s., H. sp. and H. irregulare); and (3) the lineage associated to firs and spruces (H. abietinum, H. occidentale, H. parviporum, and H. subparviporum).
Taxonomy
Heterobasidion armandii Y. C. Dai, Jia J. Chen and Yuan Yuan, sp. nov. Figures 2, 3
FIGURE 2.
Basidiocarps of Heterobasidion armandii (holotype, Dai 17605). Bar = 2 cm.
FIGURE 3.
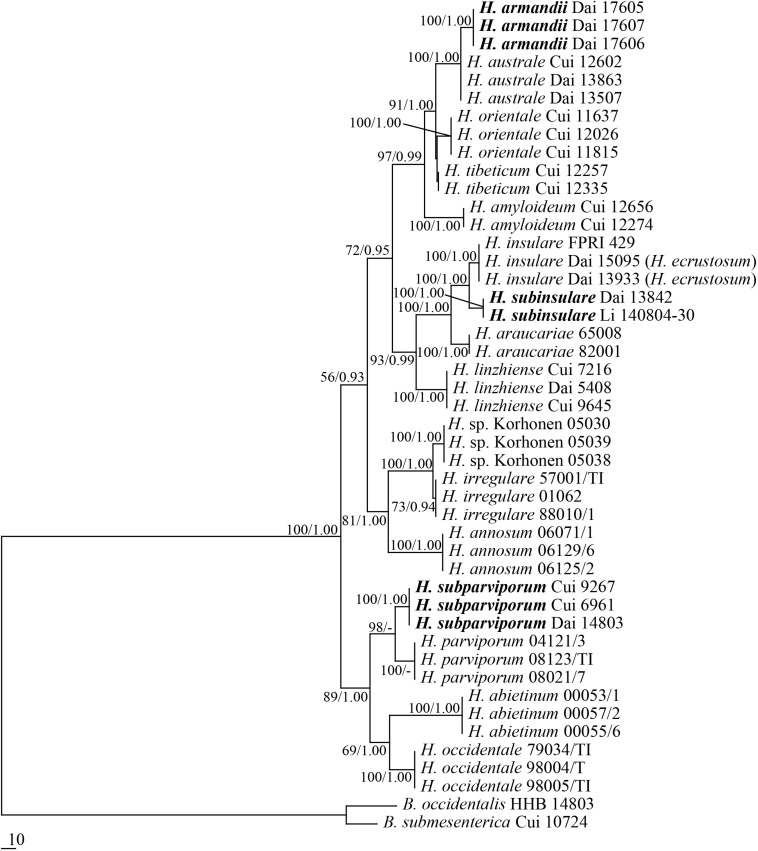
Microscopic structures of Heterobasidion armandii (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context.
MycoBank MB 834572.
Type
China, Yunnan Province, Xiping County, Mopanshan Forest Park, alt.135 m, on stump of Pinus armandii, June 15, 2017, YC Dai 17605 (BJFC025137, holotype).
Diagnosis
Differs from other Heterobasidion species by its contextual skeletal hyphae are positive in Melzer’s reagent, absence of cystidia, presence of cystidioles, and subglobose to broadly ellipsoid basidiospores measuring 4.9−5.9 × 3.9−4.5 μm.
Etymology
Armandii (Lat.): referring to the species growing on P. armandii.
Description
Basidiocarps annual, pileate, usually imbricate, leathery and without odor or taste when fresh, corky when dry. Pilei semicircular to fan-shaped, projecting up to 3 cm, 7 cm wide, and 8 mm thick at base. Pileal surface white to cream when juvenile, becoming olivaceous buff with age, at least reddish brown to dark reddish brown at base, crustose, distinctly zonate; margin cream, blunt. Pore surface white when fresh, cream when dry, not glancing; pores mostly round to angular, (3−)4−5 per mm; dissepiments thin, entire to slightly lacerate. Context cream, woody hard when dry, azonate, up to 5 mm thick, with a thin black line under crust except for the margin. Tubes cream to buff, hard corky, up to 3 mm long. Hyphal system dimitic; generative hyphae without clamp connections; tramal skeletal hyphae dextrinoid, CB+, contextual skeletal hyphae weakly IKI+, CB+; hyphae unchanged in KOH (not dissolved). Contextual generative hyphae frequently present, colorless, thin- to slightly thick-walled, frequently simple septate, occasionally branched, 3–4 mm diam; contextual skeletal hyphae dominant, colorless, thick-walled with a wide to narrow lumen, rarely branched, flexuous, interwoven, 4–5.5 mm diam. Tramal generative hyphae infrequent, hyaline, thin-walled, frequently simple septate, occasionally branched, 2–3 mm diam; tramal skeletal hyphae dominant, hyaline, thick-walled with a wide to narrow lumen, rarely branched, flexuous, strongly interwoven without orientation, 3–4 mm diam. Cystidia absent. Cystidioles present, fusiform, occasionally with an apical simple septum. Basidia clavate to ampullaceal, with a simple basal septum and four sterigmata, 14−24 × 6−8 mm. Basidioles in shape similar to basidia, but distinctly shorter. Basidiospores subglobose to broadly ellipsoid, hyaline, fairly thick-walled, asperulate, mostly bearing a small guttule, IKI−, CB+, (4.8−)4.9−5.9(−6) × (3.8−)3.9−4.5(−4.8) mm, L = 5.12 mm, W = 4.11 mm, Q = 1.24–1.25 (n = 60/2).
Additional materials (paratypes) examined
China, Yunnan Province, Xiping County, Mopanshan National Park, alt.1450 m, on stump of P. armandii, June 15, 2017, Dai 17606 (BJFC025138), Dai 17607 (BJFC025139); August 16, 2019, Dai 20410 (BJFC032078). Luquan County, Jiaozishan Forest Park, alt.2650 m, on stump of P. armandii, November 4, 2018, Dai 19258 (BJFC027726), Dai 19259 (BJFC027727), Dai 19260 (BJFC027728) and Dai 19261 (BJFC027729).
Heterobasidion subinsulare Y. C. Dai, Jia J. Chen and Yuan Yuan, sp. nov. Figures 4, 5
FIGURE 4.
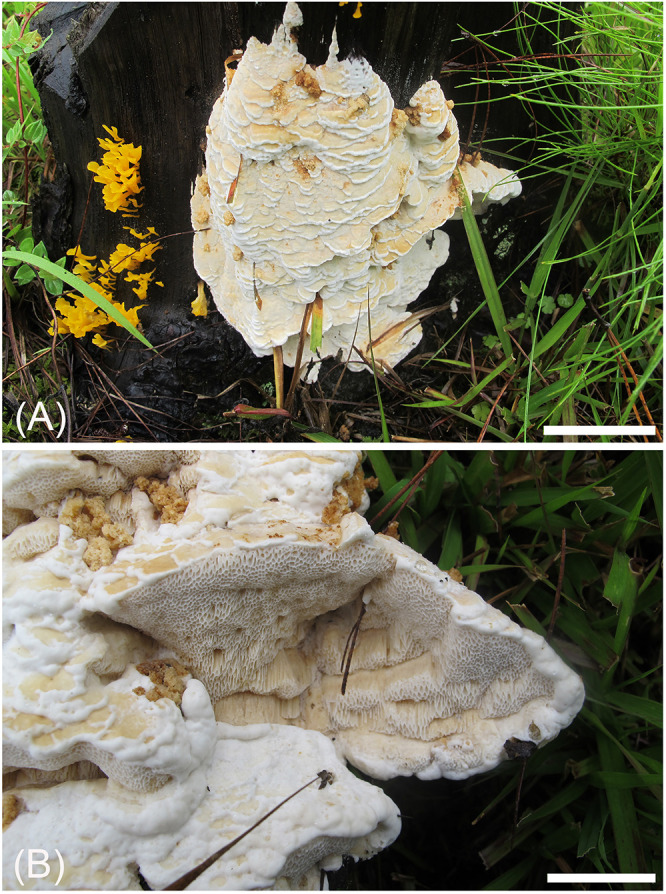
Basidiocarps of Heterobasidion subinsulare (holotype, Dai 13842). Bars (A) = 5 cm, (B) = 1 cm.
FIGURE 5.
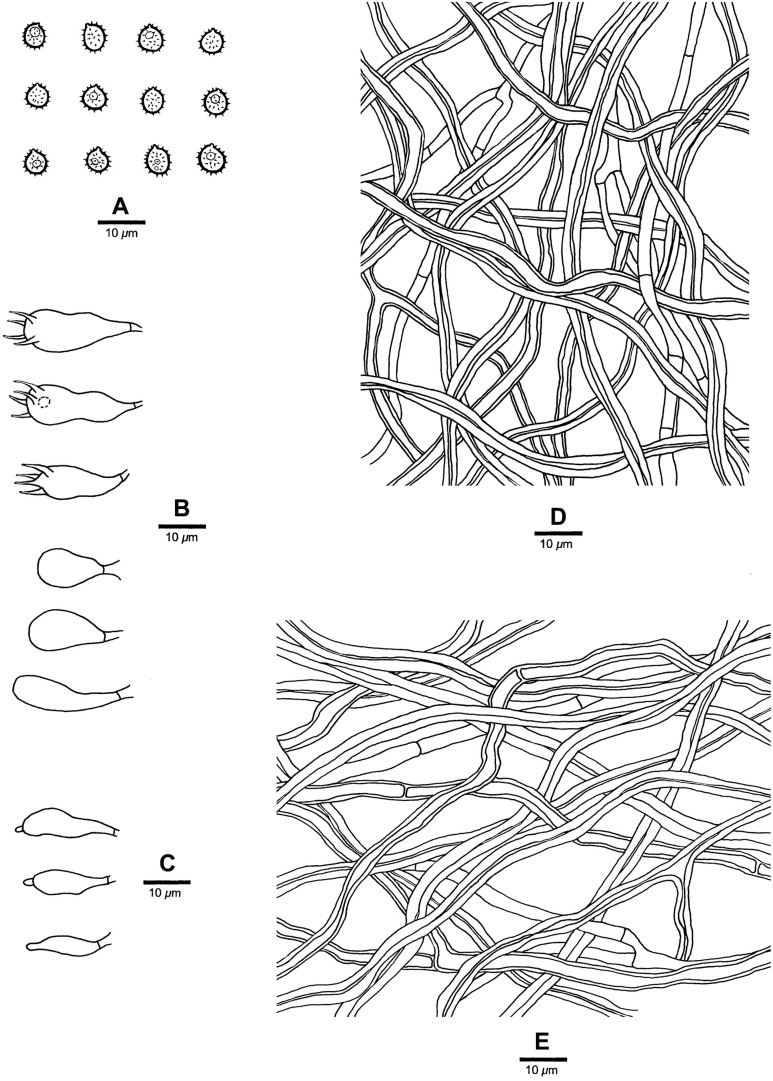
Microscopic structures of Heterobasidion subinsulare (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Cystidia; (E) Hyphae from trama; (F) Hyphae from context.
MycoBank MB 834573.
Type
China, Yunnan Province, Tengchong County, Shuanghe Village, on stump of Pinus sp., August 5, 2014, YC Dai 13842 (BJFC017572, holotype).
Diagnosis
Differs from Heterobasidion species by big pores (1–3 per mm), a non-glancing pore surface, its contextual skeletal hyphae are negative in Melzer’s reagent, presence of cystidia and cystidioles, and subglobose to broadly ellipsoid basidiospores measuring 5−5.7 × 3.8−5 μm.
Etymology
Subinsulare (Lat.): referring to the similarity to H. insulare.
Description
Basidiocarps annual, effused-reflexed to pileate, usually imbricate, leathery and without odor or taste when fresh, woody hard when dry. Pilei semicircular to fan-shaped, projecting up to 4 cm, 10 cm wide, and 4 cm thick at base. Pileal surface cream when dry, becoming buff to buff-yellow, crustose, azonate; margin buff-yellow to honey-yellow, blunt. Pore surface white when fresh, buff to clay-buff when dry, not glancing; pores angular to elongated, 1–3 per mm; dissepiments thin, entire to slightly lacerate. Context cream to buff-yellow, corky when dry, azonate, up to 0.5 cm thick. Tubes white to buff, hard corky, up to 3.5 cm long. Hyphal system dimitic; generative hyphae without clamp connections; skeletal hyphae CB+, dextrinoid near to the tube mouths, IKI− in other parts; hyphae unchanged in KOH (not dissolved). Contextual generative hyphae frequent, hyaline, thin- to slightly thick-walled, frequently simple septate and branched, 2–4 μm diam.; contextual skeletal hyphae dominant, hyaline, thick-walled with a wide lumen, rarely branched, flexuous, interwoven, 2.5–6 μm diam. Tramal generative hyphae frequent, hyaline, thin- to slightly thick-walled, occasionally simple septate, frequently branched, 2–3.5 μm diam; tramal skeletal hyphae dominant, hyaline, thick-walled with a wide lumen, rarely branched, flexuous, strongly interwoven without orientation, 2–5 μm diam. Cystidia present, thin-walled, clavate, moniliform or ventricose, 27−40 × 4−8 mm. Cystidioles present, thin-walled, fusiform, mostly with an apical simple septum, 22−25 × 4−8 μm. Basidia clavate to uniform, with a simple basal septum and four sterigmata, 18−28 × 4−6 μm. Basidioles in shape similar to basidia, but distinctly shorter. Basidiospores subglobose to broadly ellipsoid, hyaline, fairly thick-walled, asperulate, mostly bearing a small guttule, IKI−, CB+, (4.5−)5−5.7(−6) × (3.6−)3.8−5(−5.5) μm, L = 5.17 μm, W = 4.22 μm, Q = 1.22 (n = 30/1).
Additional material (paratype) examined
China, Yunnan Province, Tengchong County, on stump of Pinus sp., August 4, 2014, Li 140804-30 (BJFC018422).
Heterobasidion subparviporum Y. C. Dai, Jia J. Chen and Yuan Yuan, sp. nov. Figures 6, 7
FIGURE 6.
Basidiocarps of Heterobasidion subparviporum (paratype, Dai 14803). Bar = 4 cm.
FIGURE 7.
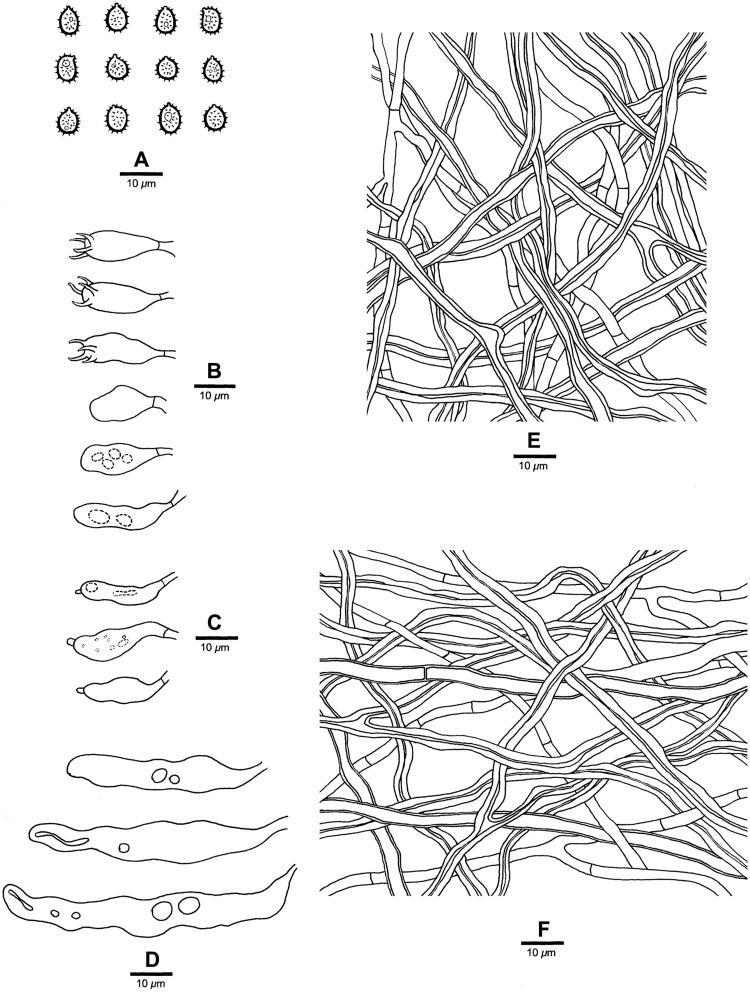
Microscopic structures of Heterobasidion subparviporum (drawn from the holotype). (A) Basidiospores; (B) Basidia and basidioles; (C) Cystidioles; (D) Hyphae from trama; (E) Hyphae from context.
MycoBank MB 834574.
Type
China, Hebei Province, Xinglong County, Wulingshan Nature Reserve, on fallen trunk of Larix sp., July 30, 2009, BK Cui 6961 (BJFC005448, holotype).
Diagnosis
Differs from Heterobasidion species by mostly round pores (3–5 per mm), amyloid contextual skeletal hyphae, absence of cystidia, presence of cystidioles, and subglobose to broadly ellipsoid basidiospores measuring 5−6.5 × 4−5.2 μm.
Etymology
Subparviporum (Lat.): referring to the similarity to H. parviporum.
Description
Basidiocarps perennial, pileate, usually imbricate, leathery and without odor or taste when fresh, hard corky when dry. Pilei semicircular to fan-shaped, projecting up to 6 cm, 9 cm wide, and 2.2 cm thick at base. Pileal surface buff to grayish brown or grayish dark, at least dark brown at base, crustose, distinctly zonate; margin cream to buff, dull, up to 2 mm. Pore surface white when fresh, cream to buff when dry, glancing; pores mostly round, occasionally irregular, 3–5 per mm; dissepiments thin, entire. Context buff to brown, corky when dry, azonate, up to 2 mm thick, with a thin black line under crust except for the margin. Tubes cream, hard corky, up to 20 mm long. Hyphal system dimitic; generative hyphae mostly simple septate; tramal skeletal hyphae dextrinoid, CB+; contextual skeletal hyphae IKI+, CB+, hyphae unchanged in KOH (not dissolved). Contextual generative hyphae infrequent, hyaline, thin-walled to slightly thick-walled, frequently simple septate and branched, 2–4 μm diam; contextual skeletal hyphae dominant, hyaline, thick-walled with a wide to narrow lumen, rarely branched, flexuous, interwoven, 2–4.5 μm diam. Tramal generative hyphae frequent, hyaline, thin-walled to slight thick-walled, frequently simple septate and branched, 1.7–3 μm diam; tramal skeletal hyphae dominant, hyaline, thick-walled with a narrow lumen, rarely branched, flexuous, strongly interwoven without orientation 1.5–3.5 μm diam. Cystidia absent. Cystidioles present, thin-walled, subulate, and ventricose, 13−26 × 4−6 μm, sometimes with a septum at the top. Basidia clavate to barrel-shaped, with a simple basal septum and four sterigmata, 18−24 × 4.5−8 μm. Basidioles in shape similar to basidia, but slightly smaller. Basidiospores subglobose to broadly ellipsoid, hyaline, fairly thick-walled, asperulate, IKI−, CB+, 5−6.5(−7) × (3.8−)4−5.2 μm, L = 5.65 μm, W = 4.35 μm, Q = 1.30–1.32 (n = 60/2).
Additional materials (paratypes) examined
China, Jilin Province, Antu County, Changbaishan Nature Reserve, on fallen trunk of Picea sp., September 13, 2014, Dai 14803 (BJFC017915); on living tree of Abies sp., September 21, 2019, Dai 20873 (BJFC032542). Xizang Autonomous Region (Tibet), Linzhi County, Lulang, on stump of Picea sp., September 16, 2010, Cui 9267 (BJFC008206).
Other materials examined
—Heterobasidion abietinum. Italy, on Abies sp., April 28, 2005, Dai 6557 (BJFC000943).
—Heterobasidion amyloideum. China, Xizang Auto. Reg. (Tibet), Linzhi County, Lulang, Sejila Mt., on fallen trunk of Abies sp., September 23, 2014, Cui 12274 (BJFC017155); Motuo County, on dead tree of Abies sp., September 21, 2014, Cui 12240 (BJFC017154); Milin County, Naligou, on fallen gymnosperm trunk, August 18, 2012, Li 1675 (isotype BJFC16026).
—Heterobasidion annosum. Belgium, on Betula sp., December 3, 2005, Dai 7445 (BJFC000949).
—Heterobasidion araucariae. New Zealand, March 20, 1985, PDD 49003 (PDD).
—Heterobasidion australe. China, Zhejiang Province, Jinan County, Tianmushan Nat. Res., stump of Pinus sp., October 15, 2004, Dai 6330 (paratype BJFC000979); Yunnan Province, Nanhua County, Dazhongshan Nature Reserve, on fallen trunk of Pinus sp., September 11, 2015, Cui 12602 (BJFC028381); Huaning County, September 6, 2013, Dai 13507 (BJFC014968); Kunming, Wild duck Lake, July 28, 2014, Dai 13863 (BJFC017593).
—Heterobasidion insulare. Philippines, Luzon, Mountain Province, on Pinus insularis, 1962, FPRI 429 (CFMR). China, Chongqing, Geleshan Forest Park, stump of Pinus sp., July 23, 2014, Dai 13933 (BJFC017663); Jiangxi Province, Anyuan County, Sanbaishan Forest Park, root of Pinus sp., December 19, 2014, Dai 15095 (BJFC018207).
—Heterobasidion irregulare. United States, on Pinus sp., 2004, JV0405/3-J (JV).
—Heterobasidion linzhiense. China, Xizang Auto. Reg. (Tibet), Linzhi County, Lulang, fallen trunk of Picea sp., September 24, 2010, Cui 9645 (holotype, BJFC008582).
—Heterobasidion occidentale. United States, on Abies sp., August 2001, JV0108/87 (JV).
—Heterobasidion orientale. China, Heilongjiang Province, Yichun, Liangshui Nature Reserve, on fallen gymnosperm trunk, August 26, 2014, Cui 11637 (BJFC016831); on fallen trunk of Pinus sp., August 28, 2014, Cui 11815 (BJFC016890); on dead tree of Picea, August 31, 2014, Cui 12026 (BJFC016964).
—Heterobasidion parviporum. Estonia, on Picea sp., May 13, 1995, Dai 1930 (BJFC001016).
—Heterobasidion sp. United States, California, Lassen National Forest, on Pinus ponderosa, March 2005, Korhonen 05030, Korhonen 05038 and Korhonen 05039 (LUKE).
—Heterobasidion tibeticum. China, Xizang Auto. Reg. (Tibet), Bomi County, Tongmai, on fallen trunk of Pinus sp., September 22, 2014, Cui 12257 (BJFC017171); Linzhi County, September 24, 2014, Cui 12335 (BJFC017249); July 31, 2004, Dai 5468 (paratype, BJFC000958).
Discussion
The current phylogeny considers that Heterobasidion species belong to three species complexes: the H. annosum F complex (previously treated as the H. annosum S group, Woodward et al., 1998), the H. annosum P complex and the H. insulare complex. The F complex of H. annosum includes four species which are mainly associated to true fir species (Abies Mill., Picea abies (L.) Karst. and Tsuga (Endl.) Carrière; Linzer et al., 2008; Dalman et al., 2010). H. subparviporum is mostly found on Picea in Asia, while H. parviporum is mostly associated to Picea in Europe and H. abietinum to Abies in Europe. H. occidentale is colonizing mostly Tsuga and Abies in western North America. The H. annosum P complex includes two taxa which mostly grow on pines: H. annosum s.s. in Eurasia, H. irregulare in North America (Linzer et al., 2008; Dalman et al., 2010). The H. insulare complex includes ten species which are associated to many species of Pinophyta (Abies, Araucaria Juss., Keteleeria Carr., Larix Mill., Picea, Pinus L., Pseudolarix Gordon, Pseudotsuga Carrière and Tsuga). H. araucariae, a species from Southern Hemisphere, is clustered into H. insulare complex, and is closely related to the species H. insulare and H. subinsulare (Figure 1).
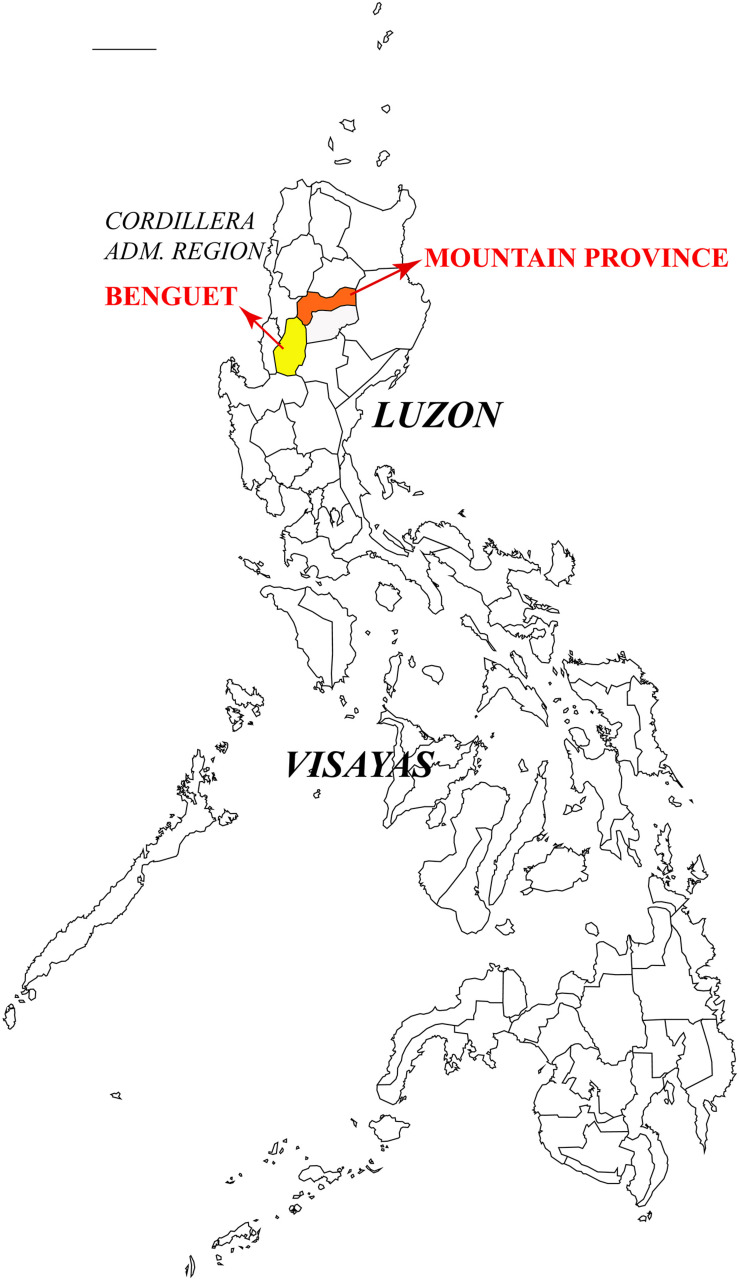
Heterobasidion insulare (=Trametes insularis Murrill) was originally described from Philippines (Murrill, 1908) and its type specimen was collected from fallen log of P. insularis in the Benguet Province, Luzon, Philippines in 1905. In 1962, Mendoza obtained the isolate FPRI-429 from P. insularis in the Mountain Province, Luzon. The Benguet Province and Mountain Province are both located in the Cordillera Administrative Region of Luzon Island (Figure 8). FPRI-429 can thus be considered as the type locality of H. insulare. The present results confirmed that FPRI-429 and representatives of H. ecrustosum are nested in the same lineage; the latter taxon was described from central Japan to Okinawa, and from southern China (Tokuda et al., 2009). We did not find any distinct morphological difference between the H. insulare type and samples of H. ecrustosum. Hence, according to the current phylogeny and morphological studies, H. ecrustosum is treated as a synonym of H. insulare.
FIGURE 8.
Type locality of Heterobasidion insulare in Philippines.
Heterobasidion armandii is closely related to H. australe (Figure 1) and the geographical distributions of the two species are overlapped in China. However, H. australe is characterized by a glancing pore surface, lacks cystidioles, and its contextual skeletal hyphae are negative in Melzer’s reagent. Morphologically, H. armandii resembles H. amyloideum and H. tibeticum by similar pores (4–6 per mm in H. amyloideum and 3–6 per mm in H. tibeticum), basidiospores (4.9−5.8 × 3.9−4.5 μm in H. amyloideum and 4.5−6 × 3.6−5.3 μm in H. tibeticum) and amyloid contextual skeletal hyphae, but H. amyloideum and H. tibeticum differ from H. armandii by the presence of cystidia (Dai and Korhonen, 2009; Chen et al., 2014).
Heterobasidion subinsulare is closely related to H. insulare (Figure 1), but the latter lacks cystidia. H. subinsulare resembles H. amyloideum and H. tibeticum by having cystidia, but the latter two species have smaller pores (3–6 per mm) and amyloid contextual skeletal hyphae (Chen et al., 2014). H. subinsulare, H. araucariae, and H. orientale share similar pores, but H. araucariae can be distinguished from H. subinsulare by longer basidiospores (5.8–6.5 μm vs. 5–5.7 μm) and lacking of cystidia, and H. orientale differs from H. subinsulare by the sharp pileal margin, dark reddish pileal surface and lacking of cystidia. In addition, H. subinsulare is distantly related to H. amyloideum, H. tibeticum, H. araucariae, and H. orientale in our current phylogeny (Figure 1).
Heterobasidion subparviporum is closely related to H. parviporum (Figure 1), and the latter was considered as same as the former according to the mating tests (Dai and Korhonen, 1999, 2003; Dai et al., 2006; Dai, 2012). Although both taxa are compatible in laboratory, they form two distinct lineages in our phylogeny (Figure 1). Morphologically, H. subparviporum differs from H. parviporum by longer cystidia (18–24 μm vs. 13–17 μm) and bigger basidiospores (5−6.5 × 4−5.2 μm vs. 4.2−5 × 3.8−4.2 μm). In addition, H. parviporum is a pathogen on P. abies in Europe, while H. subparviporum seems to be a saprophytic species according to our investigations. Based on the above data, we suggest that this Asian taxon is a new species H. subparviporum. The situation is similar with the European taxa H. parviporum and H. abietinum. These two taxa are partly sexually compatible (Capretti et al., 1990; Stenlid and Karlsson, 1991; Woodward et al., 1998), but they do not produce hybrids in nature. So they have been accepted at the species level (Niemelä and Korhonen, 1998; Otrosina and Garbelotto, 2010; Ryvarden and Melo, 2017).
Heterobasidion irregulare was proposed by Otrosina and Garbelotto (2010), and it was originally described as Polyporus irregularis Underwood on pine log from Auburn, Alabama, eastern United States (Underwood, 1897), although P. irregularis is an illegitimate name because there was earlier a fungus named P. irregularis Pers. (Persoon, 1825). The lectotype (NY730756) of H. irregulare was selected from the type material of P. irregularis Underwood, and the epitype (UC1935442) was selected from stump of P. ponderosa in the Modoc National Forest, California, western United States. However, three isolates Korhonen 05030, Korhonen 05038, and Korhonen 05039 associated to P. ponderosa from Lassen National Forest in California formed another lineage which is closely related to H. irregulare (Figure 1). Hence it is possible that another taxon exists in western North America. We did not have the basidiocarps of isolates Korhonen 05030, Korhonen 05038, and Korhonen 05039, and no information on their ecology. For the time being we treat this possible taxon as H. sp.
Heterobasidion amyloideopsis was described from Pakistan mostly based on phylogenetic analysis (Zhao et al., 2017). We studied its ITS (KT598384, KT598385), nrLSU (KT598386, KT598387), RPB1 (KT598390, KT598391), and RPB2 (KT598388, KT598389) sequences and found some of the sequences are uncorrect, and some of these sequences were deleted by NCBI. So the status of H. amyloideopsis is ambiguous.
A comparison of these three new species and their morphological and/or phylogenetically related species is also provided in Supplementary Appendix 1. The phylogenetic analyses on single loci (ITS, nrLSU, RPB1, RPB2, and GAPDH) were shown in Supplementary Figures 1–5).
Conclusion
To date, 15 species are recorded in the genus Heterobasidion, including three new species described in the present study.
Five species, H. abietinum, H. annosum s.s., H. irregulare, H. occidentale, and H. parviporum, distributed in Europe and North America are forest pathogens. Ten Asian taxa are all saprotrophs, and the Asian countries ought to consider these five European and North American species as entry plant quarantine fungi. Parallelly, European countries should consider the American H. occidentale and H. irregulare as entry plant quarantine fungi (although the latter species is already in Italy), while North America should treat H. abietinum, H. annosum s.s. and H. parviporum as entry plant quarantine fungi. Eight Heterobasidion species found in the Himalayas suggest that the ancestral Heterobasidion species may have occurred in Asia, as was proposed also in the previous divergence and biogeographic studies on the genus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YY, J-JC, and Y-CD designed the research and contributed to data analysis and interpretation. YY and J-JC performed the research. Y-CD and KK collected the materials. All authors wrote and revised the manuscript, contributed to the article, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Bao-Kai Cui (China) for allowing us to study his collections. We express our gratitude to Dr. Daniel L. Lindner (CFMR, United States) for loan of stock, to Dr. De-Wei Li (United States) for providing some literatures and improving this manuscript, and to Chang-Lin Zhao (China) for providing sequences.
Funding. The research is supported by the Second Tibetan Plateau Scientific Expedition and Research Program (STEP, No. 2019QZKK0503).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.596393/full#supplementary-material
Phylogeny of ITS.
Phylogeny of nrLSU.
Phylogeny of RPB1.
Phylogeny of RPB2.
Phylogeny of GAPDH.
A comparison of taxa in the Heterobasidion.
References
- Asiegbu F. O., Abu S., Stenlid J., Johansson M. (2004). Sequence polymorphism and molecular characterization of laccase genes of the conifer pathogen Heterobasidion annosum. Mycol. Res. 108 136–148. 10.1017/S0953756203009183 [DOI] [PubMed] [Google Scholar]
- Buchanan P. K. (1988). A new species of Heterobasidion (Polyporaceae) from Australasia. Mycotaxon 32 325–337. [Google Scholar]
- Capretti P., Korhonen K., Mugnai L., Romagnoli C. (1990). An intersterility group of Heterobasidion annosum, specialized to Abies alba. Eur. J. Plant Pathol. 20 231–240. 10.1111/j.1439-0329.1990.tb01134.x [DOI] [Google Scholar]
- Chen J. J., Cui B. K., Zhou L. W., Korhonen K., Dai Y. C. (2015). Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 71 185–200. 10.1007/s13225-014-0317-2 [DOI] [Google Scholar]
- Chen J. J., Korhonen K., Li W., Dai Y. C. (2014). Two new species of the Heterobasidion insulare complex based on morphology and molecular data. Mycoscience 55 289–298. 10.1016/j.myc.2013.11.002 [DOI] [Google Scholar]
- Dai Y. C. (2012). Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 53 49–80. 10.1007/s10267-011-0134-3 [DOI] [Google Scholar]
- Dai Y. C., Korhonen K. (1999). Heterobasidion annosum group S identified in north-eastern China. Eur. J. Plant Pathol. 29 273–279. 10.1046/j.1439-0329.1999.00153.x [DOI] [Google Scholar]
- Dai Y. C., Korhonen K. (2003). First report of Heterobasidion parviporum (S group of H. annoum sensu lato) on Tsuga spp. in Asia. Plant Dis. 87:1007. 10.1094/PDIS.2003.87.8.1007B [DOI] [PubMed] [Google Scholar]
- Dai Y. C., Korhonen K. (2009). Heterobasidion australe, a new polypore derived from the Heterobasidion insulare complex. Mycoscience 50 353–356. 10.1007/S10267-009-0491-3 [DOI] [Google Scholar]
- Dai Y. C., Vainio E. J., Hantula J., Niemelä T., Korhonen K. (2002). Sexuality and intersterility within the Heterobasidion insulare complex. Mycol. Res. 106 1435–1448. 10.1017/S0953756202006950 [DOI] [Google Scholar]
- Dai Y. C., Vainio E. J., Hantula J., Niemelä T., Korhonen K. (2003). Investigations on Heterobasidion annosum s.lat. in central and eastern Asia with the aid of mating tests and DNA fingerprinting. For. Pathol. 33 269–286. 10.1046/j.1439-0329.2003.00328.x [DOI] [Google Scholar]
- Dai Y. C., Yu C. J., Wang H. C. (2007). Polypores from eastern Xizang (Tibet), western China. Ann. Bot. Fenn. 44 135–145. [Google Scholar]
- Dai Y. C., Yuan H. S., Wei Y. L., Korhonen K. (2006). New records of Heterobasidion parviporum in China. For. Pathol. 36 287–293. 10.1111/j.1439-0329.2006.00458.x [DOI] [Google Scholar]
- Dalman K., Olson A., Stenlid J. (2010). Evolutionary history of the conifer root rot fungus Heterobasidion annosum sensu lato. Mol. Ecol. 19 4979–4993. 10.1111/j.1365-294X.2010.04873.x [DOI] [PubMed] [Google Scholar]
- Farris J. S., Källersjö M., Kluge A. G., Bult C. (1994). Testing significance of incongruence. Cladistics 10 315–319. 10.1111/j.1096-0031.1994.tb00181.x [DOI] [Google Scholar]
- Felsenstein J. (1985). Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Gilbertson R. L., Ryvarden L. (1986). North American Polypores 1. Abortiporus - Lindtneria. Oslo: Fungiflora, 1–433. [Google Scholar]
- Hall T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Huelsenbeck J. P., Bull J. J., Cunningham E. (1996). Combining data in phylogenetic analysis. Trends Ecol. Evol. 11 152–158. 10.1016/0169-5347(96)10006-9 [DOI] [PubMed] [Google Scholar]
- Johannesson H., Renvall P., Stenlid J. (2000). Taxonomy of Antrodiella inferred from morphological and molecular data. Mycol. Res. 104 92–99. 10.1017/S0953756299008953 [DOI] [Google Scholar]
- Johannesson H., Stenlid J. (2003). Molecular markers reveal genetic isolation and phylogeography of the S and F intersterility group of the wood-decay fungus Heterobasidion annosum. Mol. Phylogenet. Evol. 29 94–101. 10.1016/S1055-7903(03)00087-3 [DOI] [PubMed] [Google Scholar]
- Justo A., Hibbett D. S. (2011). Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset. Taxon 60 1567–1583. 10.1002/tax.606003 [DOI] [Google Scholar]
- Korhonen K. (1978). Intersterility groups of Heterobasidion annosum. Commun. Inst. For. Fenn. 94:25. [Google Scholar]
- Linzer R. E., Otrosina W. J., Gonthier P., Bruhn J., Laflamme G., Bussières G., et al. (2008). Inferences on the phylogeography of the fungal pathogen Heterobasidion annosum, including evidence of interspecific horizontal genetic transfer and of human-mediated, long range dispersal. Mol. Phylogenet. Evol. 46 844–862. 10.1016/j.ympev.2007.12.010 [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationship among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Maijala P., Harrington T. C., Raudaskoski M. (2003). A peroxidase gene family and gene trees in Heterobasidion and related genera. Mycologia 95 209–221. 10.2307/3762032 [DOI] [PubMed] [Google Scholar]
- Matheny P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 35 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Matheny P. B., Liu Y. J., Ammirati J. F., Hall B. D. (2002). Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 89 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- Murrill W. A. (1908). Additional philippine polyporaceae. Bull. Torrey Bot. Club 35 391–416. 10.2307/2479285 [DOI] [Google Scholar]
- Niemelä T., Korhonen K. (1998). “Taxonomy of the genus Heterobasidion,” in Heterobasidion annosum: Biology, Ecology, Impact and Control, eds Woodward S., Stenlid J., Karjalainen R., Hüttermann A. (Oxon: CAB International; ), 27–33. 10.14492/hokmj/1381758488 [DOI] [Google Scholar]
- Núñez M., Ryvarden L. (2001). East Asian polypores 2. Polyporaceae s. lato. Syn. Fung. 14 469–522. [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest 2.2. Program Distributed by the Author. Uppsala: Uppsala University. [Google Scholar]
- Ota Y., Tokuda S., Buchanan P. K., Hattori T. (2006). Phylogenetic relationships of Japanese species of Heterobasidion – H. annosum sensu lato and an undetermined Heterobasidion sp. Mycologia 98 717–725. 10.3852/mycologia.98.5.717 [DOI] [PubMed] [Google Scholar]
- Otrosina W. J., Chase T. E., Cobb F. W., Korhonen K. (1993). Population structure of Heterobasidion annosum from North America and Europe. Can. J. Bot. 71 1064–1071. 10.1139/b93-123 [DOI] [Google Scholar]
- Otrosina W. J., Garbelotto M. (2010). Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov.: a disposition of North American Heterobasidion biological species. Fungal Biol. 114 16–25. 10.1016/j.mycres.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Page R. D. M. (1996). Treeview: application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. 10.1093/bioinformatics/12.4.357 [DOI] [PubMed] [Google Scholar]
- Persoon C. H. (1825). Mycologia Europaea 2. Palmii: langen, 1–214. [Google Scholar]
- Petersen J. H. (1996). Farvekort. The Danish Mycological Society’s Color-Chart. Greve: Foreningen til Svampekundskabens Fremme. [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ryvarden L., Gilbertson R. (1993). European polypores 1. Syn. Fung. 7 1–387. [Google Scholar]
- Ryvarden L., Melo I. (2017). Poroid fungi of Europe. 2th Edition. Syn. Fung. 37 1–431. [Google Scholar]
- Stenlid J., Karlsson J. O. (1991). Partial intersterility in Heterobasidion annosum. Mycol. Res. 95 1153–1159. 10.1016/S0953-7562(09)80004-X [DOI] [Google Scholar]
- Swofford D. L. (2002). PAUP∗: Phylogenetic analysis Using Parsimony (∗and other Methods), version 4.0 Beta. Sunderland, MA: Sinauer. [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S., Hattori T., Dai Y. C., Ota Y., Buchanan P. K. (2009). Three species of Heterobasidion (Basidiomycota, Hericiales), H. parviporum, H. orientale sp. nov. and H. ecrustosum sp. nov. from East Asia. Mycoscience 50 190–202. 10.1007/S10267-008-0476-7 [DOI] [Google Scholar]
- Underwood L. M. (1897). Some new fungi, chiefly from Alabama. Bull. Torrey Bot. Club 24 81–86. 10.2307/2477799 [DOI] [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (New York, NY: Academic Press; ), 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Woodward S., Stenlid J., Karjalainen R., Hüttermann A. (1998). “Preface,” in Heterobasidion annosum: Biology, Ecology, Impact and Control, eds Woodward S., Stenlid J., Karjalainen R., Huttermann A. (Oxon: CAB International; ), xi–xii. [Google Scholar]
- Zhao C. L., Saba M., Khalid A. N., Song J., Pfister D. H. (2017). Heterobasidion amyloideopsis sp. nov. (Basidiomycota, Russulales) evidenced by morphological characteristics and phylogenetic analysis. Phytotaxa 137 199–210. 10.11646/phytotaxa.317.3.4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of ITS.
Phylogeny of nrLSU.
Phylogeny of RPB1.
Phylogeny of RPB2.
Phylogeny of GAPDH.
A comparison of taxa in the Heterobasidion.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.