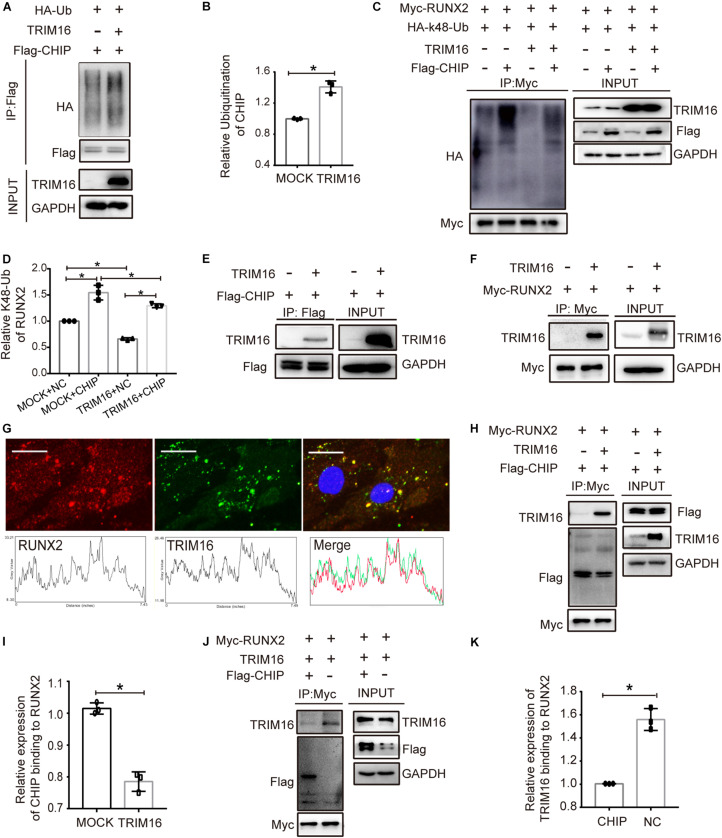
FIGURE 5.
TRIM16 decreased CHIP-mediated ubiquitination degradation of RUNX2. (A,B) Ubiquitination of CHIP influenced by TRIM16. Immunoblot analysis of lysates from cells transfected with Flag-CHIP, HA-tagged ubiquitination (HA-Ub) with or without a TRIM16 overexpression plasmid, followed by IP with anti-Flag, and probed with anti-HA (n = 3). (C,D) Ubiquitination of RUNX2 influenced by CHIP and TRIM16. Immunoblot analysis of lysates from cells transfected with Myc-RUNX2, HA-tagged K48-linked polyubiquitination (HA-k48-Ub) with or without a TRIM16 overexpression plasmid or Flag-CHIP (n = 3). (E) Coimmunoprecipitation was performed to detect the interaction of CHIP and TRIM16. Cells were transfected with Flag-CHIP, a TRIM16 overexpression plasmid, followed by IP with anti-Flag, and probed with anti-TRIM16 (n = 3). (F) Coimmunoprecipitation was performed to detect the interaction of TRIM16 and RUNX2. Cells transfected with Myc-RUNX2, a TRIM16 overexpression plasmid, followed by IP with anti-Myc, and probed with anti-TRIM16 (n = 3). (G) Colocalization analysis between TRIM16 and RUNX2 was performed in hPDLSCs. Red fluorescence represent the expression of RUNX2, green fluorescence represents the expression of TRIM16. Scale bars, 25μm. (H,I) Coimmunoprecipitation was performed to detect the interaction of CHIP and RUNX2 influenced by TRIM16, followed by IP with anti-Myc, and probed with anti-Flag (n = 3). (J,K) Coimmunoprecipitation was performed to detect the interaction of TRIM16 and RUNX2 influenced by CHIP, followed by IP with anti-Myc, and probed with anti-TRIM16 (n = 3). *P < 0.05 vs. the control group, respectively.