Abstract
Maintenance of cellular homeostasis and genome integrity is a critical responsibility of DNA double-strand break (DSB) signaling. P53-binding protein 1 (53BP1) plays a critical role in coordinating the DSB repair pathway choice and promotes the non-homologous end-joining (NHEJ)-mediated DSB repair pathway that rejoins DSB ends. New insights have been gained into a basic molecular mechanism that is involved in 53BP1 recruitment to the DNA lesion and how 53BP1 then recruits the DNA break-responsive effectors that promote NHEJ-mediated DSB repair while inhibiting homologous recombination (HR) signaling. This review focuses on the up- and downstream pathways of 53BP1 and how 53BP1 promotes NHEJ-mediated DSB repair, which in turn promotes the sensitivity of poly(ADP-ribose) polymerase inhibitor (PARPi) in BRCA1-deficient cancers and consequently provides an avenue for improving cancer therapy strategies.
Keywords: P53-binding protein 1 (53BP1), DNA double-strand break (DSB), Non-homologous end-joining (NHEJ), Homologous recombination (HR), Poly(ADP-ribose) polymerase inhibitor (PARPi)
1 Introduction
Preserving genomic integrity and maintaining cellular homeostasis are crucial functions of DNA double-strand break (DSB) repair. Any unrepaired DSBs can trigger cell growth arrest and cell death, and unrepaired DSBs are potential inducers of oncogene activation and loss of tumor suppressors. Two mechanistically distinct pathways are involved in the elimination of DSBs from the genome: homologous recombination (HR), which requires an identical DNA template in the sister chromatid for DNA repair (Heyer et al., 2010), and non-homologous DNA end-joining (NHEJ), which rejoins the broken ends without the use of extensive homology (Lieber, 2010). Fully understanding how DNA repair pathway choice occurs is critical for designing better therapies for cancer patients. Poly(ADP-ribose) polymerase inhibitor (PARPi) is commonly used in patients with breast cancer type 1 susceptibility protein (Brca1) or Brca2 mutations (Bryant et al., 2005; Farmer et al., 2005). Surprisingly, two research groups independently found that loss of p53-binding protein 1 (53bp1) in the background of Brca1 deficiency could restore HR repair signaling and render Brca1-deficient tumors resistant to PARP inhibition treatment, which has made resistance to PARPi therapy a major problem in clinical practice (Bouwman et al., 2010; Bunting et al., 2010).
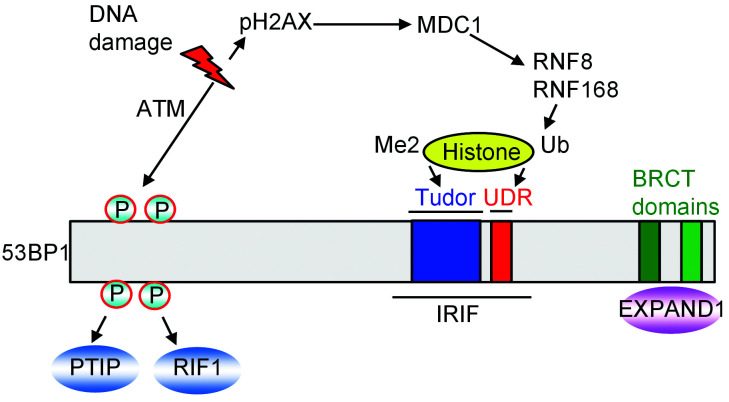
For more than two decades, 53BP1 has been described as an important regulator of DSB signaling. Studies have revealed that although 53BP1 lacks any enzymatic activity, it can recruit other responsive proteins to DNA damage sites to facilitate the NHEJ repair process. In response to DSBs, 53BP1 rapidly accumulates on the chromatin surrounding the DNA damage site (Schultz et al., 2000; Anderson et al., 2001; Rappold et al., 2001). This accumulation is driven by a signaling cascade that originates with ataxia-telangiectasia mutated (ATM) kinase-mediated phosphorylation of H2AX (known as γH2AX). γH2AX binds directly to the mediator of DNA damage checkpoint protein 1 (MDC1) at the break site and then activates RING finger protein 8 (RNF8)-RNF168-mediated ubiquitination of chromatin (Bekker-Jensen and Mailand, 2011; Lukas et al., 2011), which induces the 53BP1 recruitment to DNA lesions. The E3 ubiquitin ligases RNF168 and RNF8 collaborate with ubiquitin-conjugating enzyme E2N (UBC13), an E2-conjugating enzyme, and with other E2 enzymes to ubiquitinate the chromatin around the DNA lesion (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007; Doil et al., 2009; Stewart et al., 2009). H2A is a key substrate of RNF168, which ubiquitinates H2A on its Lys13 and/or Lys15 in response to DNA damage (Gatti et al., 2012; Mattiroli et al., 2012). RNF168 and RNF8 are at the spearhead of a complex and dynamic ubiquitination-dependent signaling cascade that has contact with almost every DSB responsive phase, including damage checkpoint activation, chromatin remodeling, transcriptional silencing, and DNA repair (Goodarzi and Jeggo, 2013).
The ionizing radiation-induced foci (IRIF) region of 53BP1, necessary for recruitment of 53BP1 to DSBs, contains the ubiquitin-dependent recruitment (UDR) motif, a tandem Tudor domain, and an oligomerization domain (Iwabuchi et al., 2003; Charier et al., 2004; Fradet-Turcotte et al., 2013). The Tudor domain binds H4K20me2 while the UDR motif binds H2AK15Ub. It has been demonstrated that the roles of 53BP1 in DNA repair depend on its accumulation in DNA damage sites via recognition of H4K20me2 (Botuyan et al., 2006; Fradet-Turcotte et al., 2013).
In this review, we highlight 53BP1 as a key determinant of DSB repair pathway choice. In particular, we focus on discoveries that have shed light on how post-transcriptional modifications (PTMs) and upstream factors regulate 53BP1 in the DNA damage response and how 53BP1 induces the downstream responsive effectors involved in the NHEJ signaling pathways.
2 53BP1 structure and interaction partners
53BP1 is a large protein of 1972 amino acids and with more than a 200-kD mass which is trans-lated by the TP53BP1 gene (Adams and Carpenter, 2006; Panier and Boulton, 2014). Although 53BP1 does not show any enzymatic activity, it consists of multiple interaction surfaces for DSB-responsive proteins. Important structural regions of 53BP1 consist of tandem Tudor domains, a dynein 8-kD light chain binding motif, a glycine-arginine-rich stretch, two BRCA1 carboxyl-terminal (BRCT) domains, and 28 amino-terminal Ser/Thr-Gln sites, which are phosphorylated by ATM kinase (Adams and Carpenter, 2006).
Three proteins that rely on 53BP1 for their recruitments to DNA damage sites are EXPAND1 (known as mutated melanoma-associated antigen 1 (MUM1)), which binds to the BRCT domains of 53BP1 in a phosphorylation-independent manner (Huen et al., 2010), and replication timing regulatory factor 1 (RIF1) and Pax transactivation domain-interacting protein (PTIP), which interact with the 53BP1 N-terminal Ser/Thr-Gln sites in an ATM-dependent manner (Munoz et al., 2007; Chapman et al., 2013) (Fig. 1).
Fig. 1. Regulation and binding proteins of 53BP1. 53BP1: p53-binding protein 1; ATM: ataxia-telangiectasia mutated; BRCT: BRCA1 carboxyl-terminal; IRIF: ionizing radiarion-induced foci; MDC1: mediator of DNA damage checkpoint protein 1; P: phosphorylated; PTIP: Pax transactivation domain-interacting protein; pH2AX: phospho-histone H2AX; RNF: RING finger protein; RIF1: regulatory factor 1; Ub: ubiquitin; UDR: ubiquitination-dependent recruitment motif.
3 Roles of 53BP1 in DSB repair pathway choice
Several milestone studies have shown that 53BP1 plays a determinant role in DSB repair pathway choice and in improving the fidelity of DSB repair (Heyer et al., 2010; Lieber, 2010; Chapman et al., 2012a). The repair pathway selection decision is crucial for maintaining genome integrity and is precisely regulated during different cell cycle phases (Chapman et al., 2012b).
3.1. 53BP1-mediated NHEJ pathway
As an important component involved in cellular response to DNA damage, 53BP1 participates in DNA damage checkpoint control and DNA repair (Abraham, 2002; Fitzgerald et al., 2009; van Gent, 2009). Subsequent studies have indicated that a loss of 53bp1 restores HR repair resulting in rescuing embryonic lethality in Brca1-knockout mice (Bouwman et al., 2010; Bunting et al., 2010). Moreover, loss of 53bp1 renders Brca1-deficient cells resistant to PARPi (Bouwman et al., 2010; Bunting et al., 2010). These significant observations suggest that 53BP1 may counteract HR repair in BRCA1-deficient cells. The current hypothesis is that BRCA1- and 53BP1-dependent pathways compete with each other during the early steps of HR repair, especially at DNA end resection. While BRCA1 promotes DNA end resection, 53BP1 may suppress end resection and therefore promote NHEJ and prevent HR repair. However, 53BP1 is an adaptor protein that recruits other proteins to sites of DNA breaks to facilitate NHEJ repair, which competes with BRCA1-dependent HR-mediated DNA repair. In the past few years, 53BP1-dependent DNA repair pathway was well established, which consists of two downstream sub-pathways that are respectively mediated by RIF1-REV7 and PTIP-Artemis (Callen et al., 2013; Chapman et al., 2013; di Virgilio et al., 2013; Escribano-Díaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013; Wang et al., 2014; Boersma et al., 2015; Xu et al., 2015). Very recently, several groups reported that the Shieldin complex acts as the downstream effector of 53BP1/RIF1/REV7 to restrict DSB resection and counteract HR (Dev et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018; Tomida et al., 2018). These two sub-pathways may have overlapping functions, but both are involved in the suppression of HR repair in BRCA1-deficient cells.
3.2. BRCA1-mediated HR pathway
When DSBs occur, a process termed DNA end resection is activated. End resection produces extensive 3' single-strand overhang, which is a key step in determining repair pathway choice, since ends that bear long 3' DNA tails are destined for HR repair, while at the same time such ends are not substrates for NHEJ and therefore would prevent NHEJ repair. BRCA1 promotes DNA end resection and mediates the HR repair pathway.
A critical question is how BRCA1 overcomes the barrier to resection that is enforced by 53BP1. 53BP1 is phosphorylated by ATM kinase in the S phase, which promotes RIF1 recruitment at DSBs and inhibits DNA end-resection. Isono et al. (2017) have shown that BRCA1 promotes 53BP1 dephosphorylation by PP4C phosphatase and RIF1 release, thus facilitating end resection to mediate HR repair. 53BP1 acetylation has also been implicated in the choice of DNA repair pathways. CREB-binding protein (CBP)-mediated acetylation of 53BP1 inhibits the recruitment of 53BP1 and its downstream factors PTIP and RIF1 to DSBs, which suppresses NHEJ repair and promotes BRCA1-mediated HR repair (Guo et al., 2018). The meiotic recombination 11 (MRE11)/DNA repair protein RAD50 homolog (RAD50)/Nijmegen breakage syndrome 1 (NBS1) (MRN)-CTBP-interacting protein (CtIP) pathway may potentially promote HR repair in a BRCA1-dependent manner. CtIP has been shown to inhibit RIF1 end resection blocking activity in the S/G2 phase (Escribano-Díaz et al., 2013; Zimmermann et al., 2013). Recent studies demonstrate that CtIP promotes end resection and mediates the rescue of genomic stability of mouse Brca1/53bp1-deficient cells (Polato et al., 2014).
4 Upstream regulators of 53BP1 in DSB signaling
As described above, 53BP1 is recruited to the DNA damage sites, a process depends on the Tudor domain of 53BP1 recognizing the lysine methylation of histone.
4.1. 53BP1 recruitment to chromatin regulated by Tudor-interacting repair regulator (TIRR)
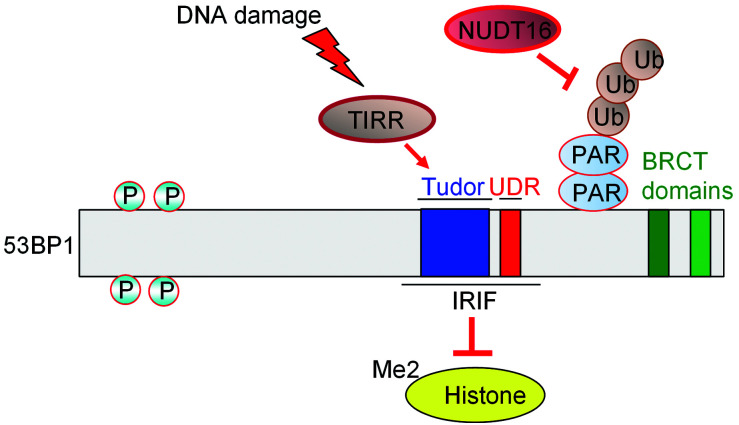
A new protein was recently identified in the 53BP1 signaling pathway (Drané et al., 2017; Zhang et al., 2017; Botuyan et al., 2018; Dai et al., 2018; Wang et al., 2018). NUDT16L1, now known as the TIRR, which is a member of the family of nucleoside diphosphate-linked moiety X (Nudix) hydrolases, was shown to interact with and regulate the activity of the IRIF region of 53BP1 (Drané et al., 2017; Zhang et al., 2017). At a DSB, the DNA ends are marked with histones H4K20me2 and H2AK15ub, which are specific targets of 53BP1. The unique function of TIRR is that it can directly bind the tandem Tudor domain of 53BP1, which prevents the binding of 53BP1 to H4K20me. Moreover, upon DNA damage, ATM phosphorylates 53BP1 and recruits RIF1 to dissociate the 53BP1-TIRR complex from chromatin, which allows 53BP1 to recognize H4K20me2 and direct the repair path choice toward the NHEJ pathway. Further observations support the finding that the interaction between 53BP1 and TIRR is of great importance for the activation of HR in BRCA1-deficient cells, which provides opportunities for developing novel chemotherapeutics (Wang et al., 2018). X-ray crystallography of TIRR-53BP1 reveals that a TIRR arginine (Arg107) residue can mask 53BP1's histone methyllysine-binding surface (Drané et al., 2017; Botuyan et al., 2018). The TIRR amino-terminal region combined with the TIRR L8-loop can prevent the methylation reader joining surface in Tudor of 53BP1, which in turn abolishes 53BP1 recruitment to nucleosomes bearing H4K20me2 (Dai et al., 2018). Further, because TIRR is an RNA-binding protein and non-coding RNAs have been implicated in the recruitment of 53BP1 to DSBs, researchers have wondered whether RNA molecules formed in the DNA damage response can disassemble the 53BPl-TIRR complex. X-ray crystallography and biochemistry assay have demonstrated that RNA molecules do disassemble the 53BP1-TIRR complex, which indicates that RNA molecules could act as a trigger for 53BP1 recruitment to chromatin in response to DNA damage (Botuyan et al., 2018).
In summary, on the one hand, TIRR stabilizes the 53BP1 protein level; depletion of TIRR decreases nuclear-soluble 53BP1 and weakens the association of 53BP1 with its binding partners (Zhang et al., 2017). On the other hand, overexpression of TIRR eliminates the association of H4K20me2 and 53BP1, which consequently suppresses the recruitment of 53BP1 to DSBs, thus disrupting 53BP1-dependent DNA repair (Drané et al., 2017; Botuyan et al., 2018).
4.2. Poly-ADP ribosylation of 53BP1 regulated by NUDT16
Recently, researchers have identified a novel post-translational modification of 53BP1 by ADP-ribosylation that is driven by the Nudix enzyme, NUDT16. NUDT16 has been shown to have in vitrohydrolase activity that removes protein ADP-ribosylation (Palazzo et al., 2015). Nudix proteins are characterized by a highly conserved 23-amino acid Nudix motif, GX 5 EX 7 REUXEEXGU, where U is an aliphatic or hydrophobic residue (McLennan, 2006). Zhang et al.(2020) have found that NUDT16 regulates the poly-ADP ribosylation of 53BP1, which is targeted by the poly(ADP-ribose) (PAR)-binding E3 ubiquitin ligase, RNF146, leading to 53BP1 poly-ubiquitination and degradation. Overexpression of a catalytically inactive NUDT16 mutant prevented 53BP1 recruitment to DSBs. Because mutant NUDT16 binding of TIRR increased after DNA damage, TIRR/53BP1 Tudor binding was enhanced, thus impairing interaction of the 53BP1 Tudor domain with H4K20me2 (Zhang et al., 2020).
TIRR and NUDT16 both belong to the Nudix hydrolase family and share a 46% sequence identity with each other. However, TIRR lacks an intact Nudix domain, which is essential for NUDT16 hydrolysis activity. Structural comparisons reveal that a TIRR histidine (H106) essential for TIRR-53BP1 Tudor binding is absent from NUDT16 (Dai et al., 2018; Wang et al., 2018). The specificity of 53BP1 Tudor recognition by TIRR is unlikely to be conferred by the Nudix domain because this domain is located far from the binding interface of TIRR/53BP1 Tudor.
Although these two upstream regulators of 53BP1 stabilize the 53BP1 protein, their mechanisms for regulating 53BP1 in the DNA damage response differ. Firstly, NUDT16, but not TIRR, shows hydrolase activity that could remove ADP-ribosylation from 53BP1, which protects 53BP1 from ADP-ribosylation-dependent ubiquitination and degradation. Secondly, TIRR overexpression inhibits 53BP1 localization to DSBs by blocking its H4K20me2-binding sites, whereas NUDT16 does not affect 53BP1 foci formation after irradiation. NUDT16 also lacks the binding residue critical for binding with 53BP1, which facilitates TIRR directly binding with the Tudor domain of 53BP1 (Zhang et al., 2020) (Fig. 2).
Fig. 2. Regulation of 53BP1 by TIRR and NUDT16. 53BP1: p53-binding protein 1; BRCT: BRCA1 carboxyl-terminal; IRIF: ionizing radiation-induced foci; NUDT16: nudix hydrolase 16; P: phosphorylated; PAR: poly(ADP-ribose); TIRR: Tudor-interacting repair regulator; Ub: ubiquitin; UDR: ubiquitin-dependent recruitment.
5 Downstream effectors of 53BP1 in the NHEJ pathway
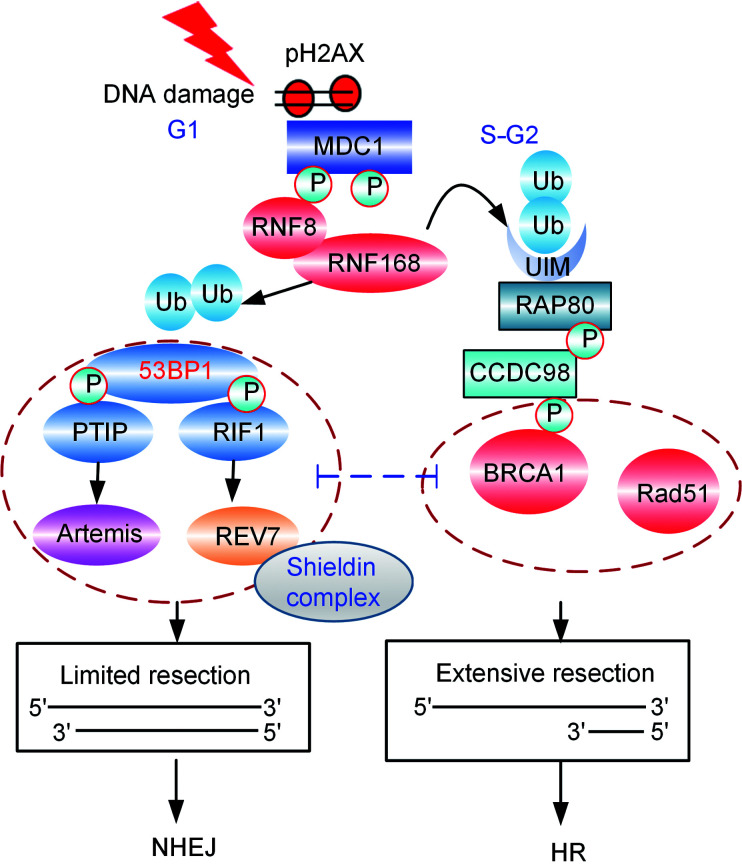
53BP1 also regulates DSB repair by suppressing the nucleolytic resection of DNA termini, which controls two downstream sub-pathways that are regulated by the RIF1-REV7/Shieldin complex and PTIP-Artemis, respectively (Fig. 3).
Fig. 3. Regulation of double-strand break repair pathway choice. 53BP1: p53-binding protein 1; BRCA1: breast cancer type 1 susceptibility protein; CCDC98: coiled-coil domain-containing protein 98; HR: homologous recombination; MDC1: mediator of DNA damage checkpoint protein 1; NHEJ: non-homologous end-joining; pH2AX: phospho-histone H2AX; P: phosphorylated; PTIP: Pax transactivation domain-interacting protein; RAP80: receptor associated protein 80; RIF1: replication timing regulatory factor 1; RNF: RING finger protein; Ub: ubiquitin; UIM: ubiquitin-interacting motif.
5.1. Cooperation between 53BP1 and RIF1-REV7/Shieldin complex in NHEJ repair
A major step forward in our understanding of how 53BP1 promotes NHEJ signaling came with the discovery that 53BP1 cooperates with RIF1 to promote NHEJ and inhibit HR repair signaling (Chapman et al., 2013; di Virgilio et al., 2013; Escribano-Díaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013).
Several observations have shown that RIF1 binds to 53BP1 directly in a DNA damage-dependent manner (di Virgilio et al., 2013). 53BP1 coimmunoprecipitates with RIF1 and recruits RIF1 to the DNA damaged sites by its N-terminal domain, which contains multiple ATM-phosphorylated Ser/ThrGln sites (Chapman et al., 2013; di Virgilio et al., 2013; Escribano-Díaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). Furthermore, similarly to the conditional ablation of 53BP1, conditional ablation of mouse Rif1 specifically in B cells resulted in a profound defect in class switch recombination (Manis et al., 2004; Ward et al., 2004; Chapman et al., 2013).
Although the NHEJ of dysfunctional telomeres is abolished in cells lacking 53BP1 and in cells expressing 53BP128A (an allele harboring alanine substitutions in all 28 N-terminal ATM/ATM and Rad3-related (ATR) sites) (Lottersberger et al., 2013), loss of RIF1 exerts a considerably milder defect. Moreover, although the generation of toxic radial chromosomes in Brca1-deficient cells is inhibited in 53bp1 -/- or in 53BP128A-mutant cells, the loss of RIF1 only partially rescues HR repair in BRCA1-deficient cells (Bouwman et al., 2010; Bunting et al., 2010, 2012; Bothmer et al., 2011).
The function of 53BP1 requires interactions with PTIP (Munoz et al., 2007) and RIF1, the latter of which recruits REV7 (also known as MAD2L2) to DSB sites (Boersma et al., 2015; Xu et al., 2015). Recently, in several persuasive studies, a number of groups have identified a novel protein complex, named Shieldin, that is a downstream effector of 53BP1-RIF1, which protects DNA ends from resection and facilitates the 53BP1-dependent NHEJ repair pathway (Dev et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018). Shieldin, a four-subunit single-stranded DNA (ssDNA)-binding complex, consists of SHLD1 (known as RINN3 or C20ORF196), SHLD2 (known as RINN2 or FAM35A), SHLD3 (known as RINN1 or CTC-534A2.2), and REV7. Among these subunits, SHLD3 is the most apical component, bridging REV7 to RIF1 and filling the gap in the 53BP1-RIF1-REV7 axis (Gupta et al., 2018; Noordermeer et al., 2018; Setiaputra and Durocher, 2019). Because of this, REV7 and SHLD3 form a subcomplex that is required for the DSB recruitment unit whereas SHLD1, along with SHLD2, acts as the ssDNA-binding module within the complex. 53BP1-RIF1-Shieldin regulates the 3' overhangs through the CTC1-STN1-TEN1 (CST) complex, which functions as an accessory factor for polymerase-α-primase, and is a downstream effector in the 53BP1 pathway. CST-Pola-mediated fill-in helps to regulate DSB repair by the 53BP1-RIF1-Shieldin axis (Mirman et al., 2018).
Further, crystal structure analysis of REV7 in complex with the REV7-binding domain of SHLD3 has revealed that SHLD3 binds with REV7 via the SHLD3 N-terminal loop and C-terminal helix, which provides critical insight into how SHLD3 recognizes REV7 (Dai et al., 2020). Taken together, 53BP1-RIF1 recruits the Shieldin complex to DNA damage sites and mediates NHEJ-related repair of immunoglobulin class-switch recombination, fusion of unprotected telomeres and intrachromosomal breaks. The Shieldin complex acts as a downstream effector of 53BP1-RIF1 to limit DNA end resection and cause BRCA1-deficient cells to be sensitive to PARPi.
5.2. PTIP-Artemis complex mediates 53BP1-dependent DNA repair
As described above, RIF1 is one of the critical factors acting as a downstream effector of 53BP1, and it antagonizes the BRCA1 function in DNA repair. However, the level of resection rescued by RIF1 loss is insufficient to rescue the HR defect in BRCA1-deficient cells. Callen et al. (2013) have revealed that depletion of PTIP provides additional or sustained end resection that is required for rescuing HR repair in BRCA1-deficient cells, which indicates that RIF1 and PTIP may act together and/or inhibit different resection steps during HR repair.
PTIP is a ubiquitously expressed nuclear protein that individualistically regulates transcription as part of the MLL3-MLL4 methyltransferase complex that catalyzes H3K4me3 (Cho et al., 2007; Munoz et al., 2007). In addition to its well-known function in transcription initiation, a separate PTIP pool performs an unknown capacity in the DNA damage response (Gong et al., 2009). Indeed, PTIP has been described in both HR (Wang et al., 2010) and NHEJ (Callen et al., 2012). PTIP contains BRCT repeats that directly interact with phosphorylated Ser25 of 53BP1 in an ATM-dependent manner (Manke et al., 2003; Yu et al., 2003; Munoz et al., 2007). Unlike RIF1, PTIP is dispensable for NHEJ during class-switch recombination, but the loss of PTIP represses toxic NHEJ-mediated DSB repair in BRCA1-defiecient cells and restores RAD51 foci (Callen et al., 2013).
PTIP participates in NHEJ repair and in preventing HR repair by recruiting a bona fide NHEJ protein, Artemis, to sites of DNA damage (Wang et al., 2014). Artemis is an evolutionally conserved nuclease that possesses endonuclease activity (Ma et al., 2002). Artemis is a component of the NHEJ pathway, which directly binds to PTIP and accumulates at DNA damage sites (Wang et al., 2014). The interaction of Artemis and PTIP is induced by DNA damage and phosphorylation dependence. Its endonuclease activity permits Artemis to trim DNA ends to promote NHEJ and, consequently, at the same time to prevent end resection and RAD51-dependent HR repair (Wang et al., 2014).
6 Concluding remarks
DNA damage induces a serious threat to genome integrity, and therefore cells have evolved elegant DNA damage response mechanisms to protect their genome stability.
The choice of repair pathways is a crucial step during DSB processing that involves several components such as epigenetic alterations, cell cycle phases, and the DNA end resection. 53BP1, in conjunction with its upstream regulators and downstream effectors, spearheads the restraints of end resection to antagonize BRCA1-mediated HR repair, and consequently promote the NHEJ pathway.
Deeper knowledge of up- and downstream regulations of 53BP1 will improve our understanding of how the 53BP1 repair pathway operates in the cell to facilitate DNA repair and through it, mechanisms of cancer-associated PARPi resistance. In the future, this direction of research will offer new targets for improving cancer therapy and overcome therapeutic resistance.
Author contributions
Fan ZHANG wrote and edited the manuscript and drew the figures. Zihua GONG contributed to the revision of the manuscript and the figures. Both authors have read and approved the final manuscript and, therefore, take responsibility for the integrity and security of the manuscript.
Compliance with ethics guidelines
Fan ZHANG and Zihua GONG declare that they have no conflict of interest.
This paper does not contain any studies with human or animal subjects performed by either of the authors.
References
- Abraham RT, 2002. Checkpoint signalling: focusing on 53BP1. Nat Cell Biol, 4(12): E277-E279. 10.1038/ncb1202-e277 [DOI] [PubMed] [Google Scholar]
- Adams MM, Carpenter PB, 2006. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div, 1: 19 10.1186/1747-1028-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y, 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol, 21(5): 1719-1729. 10.1128/MCB.21.5.1719-1729.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N, 2011. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett, 585(18): 2914-2919. 10.1016/j.febslet.2011.05.056 [DOI] [PubMed] [Google Scholar]
- Boersma V, Moatti N, Segura-Bayona S, et al. , 2015. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5' end resection. Nature, 521(7553): 537-540. 10.1038/nature14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, di Virgilio M, et al. , 2011. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell, 42(3): 319-329. 10.1016/j.molcel.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, et al. , 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell, 127(7): 1361-1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Cui GF, Drané P, et al. , 2018. Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein. Nat Struct Mol Biol, 25(7): 591-600. 10.1038/s41594-018-0083-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, et al. , 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol, 17(6): 688-695. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, et al. , 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature, 434(7035): 913-917. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, et al. , 2010. 53BP1 inhibits homologous recombination in BRCA1-deficient cells by blocking resection of DNA breaks. Cell, 141(2): 243-254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Kozak ML, et al. , 2012. Brca1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell, 46(2): 125-135. 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Faryabi RB, Luckey M, et al. , 2012. The DNA damage- and transcription-associated protein Paxip1 controls thymocyte development and emigration. Immunity, 37(6): 971-985. 10.1016/j.immuni.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, di Virgilio M, Kruhlak MJ, et al. , 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell, 153(6): 1266-1280. 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, et al. , 2012a. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci, 125(Pt 15): 3529-3534. 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MRG, Boulton SJ, 2012b. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell, 47(4): 497-510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, et al. , 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell, 49(5): 858-871. 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charier G, Couprie J, Alpha-Bazin B, et al. , 2004. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure, 12(9): 1551-1562. 10.1016/j.str.2004.06.014 [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong SH, et al. , 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4: methyltransferase complex. J Biol Chem, 282(28): 20395-20406. 10.1074/jbc.M701574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YX, Zhang AL, Shan S, et al. , 2018. Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR. Nat Commun, 9: 2123 10.1038/s41467-018-04557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YX, Zhang F, Wang LG, et al. , 2020. Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair. J Biol Chem, 295(1): 250-262. 10.1074/jbc.RA119.011464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev H, Chiang TWW, Lescale C, et al. , 2018. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol, 20(8): 954-965. 10.1038/s41556-018-0140-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Virgilio M, Callen E, Yamane A, et al. , 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science, 339(6120): 711-715. 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, et al. , 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell, 136(3): 435-446. 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Drané P, Brault ME, Cui GF, et al. , 2017. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature, 543(7644): 211-216. 10.1038/nature21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, et al. , 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell, 49(5): 872-883. 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, et al. , 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature, 434(7035): 917-921. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang JD, et al. , 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem, 288(16): 11135-11143. 10.1074/jbc.M113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald JE, Grenon M, Lowndes NF, 2009. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans, 37(Pt 4): 897-904. 10.1042/BST0370897 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Díaz C, et al. , 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature, 499(7456): 50-54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Pinato S, Maspero E, et al. , 2012. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle, 11(13): 2538-2544. 10.4161/cc.20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Oliveira C, Becker JR, et al. , 2018. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature, 560(7716): 122-127. 10.1038/s41586-018-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong ZH, Cho YW, Kim JE, et al. , 2009. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem, 284(11): 7284-7293. 10.1074/jbc.M809158200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo PA, 2013. The repair and signaling responses to DNA double-strand breaks. Adv Genet, 82: 1-45. 10.1016/B978-0-12-407676-1.00001-9 [DOI] [PubMed] [Google Scholar]
- Guo X, Bai YT, Zhao MM, et al. , 2018. Acetylation of 53BP1 dictates the DNA double strand break repair pathway. Nucleic Acids Res, 46(2): 689-703. 10.1093/nar/gkx1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Somyajit K, Narita T, et al. , 2018. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell, 173(4): 972-988.. E923 10.1016/j.cell.2018.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J, 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet, 44: 113-139. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, et al. , 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell, 131(5): 901-914. 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MSY, Huang J, Leung JWC, et al. , 2010. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell, 37(6): 854-864. 10.1016/j.molcel.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono M, Niimi A, Oike T, et al. , 2017. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep, 18(2): 520-532. 10.1016/j.celrep.2016.12.042 [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Basu BP, Kysela B, et al. , 2003. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J Biol Chem, 278(38): 36487-36495. 10.1074/jbc.M304066200 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, et al. , 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science, 318(5856): 1637-1640. 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem, 79: 181-211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger F, Bothmer A, Robbiani DF, et al. , 2013. Role of 53BP1 oligomerization in regulating double-strand break repair. Proc Natl Acad Sci USA, 110(6): 2146-2151. 10.1073/pnas.1222617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J, 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol, 13(10): 1161-1169. 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Ma YM, Pannicke U, Schwarz K, et al. , 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell, 108(6): 781-794. 10.1016/s0092-8674(02)00671-2 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, et al. , 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell, 131(5): 887-900. 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia ZF, et al. , 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol, 5(5): 481-487. 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, et al. , 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science, 302(5645): 636-639. 10.1126/science.1088877 [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JHA, van Dijk WJ, et al. , 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell, 150(6): 1182-1195. 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- McLennan AG, 2006. The Nudix hydrolase superfamily. Cell Mol Life Sci, 63(2): 123-143. 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, et al. , 2018. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature, 560(7716): 112-116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Jowsey PA, Toth R, et al. , 2007. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res, 35(16): 5312-5322. 10.1093/nar/gkm493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer SM, Adam S, Setiaputra D, et al. , 2018. The shieldin complex mediates 53BP1-dependent DNA repair. Nature, 560(7716): 117-121. 10.1038/s41586-018-0340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Thomas B, Jemth AS, et al. , 2015. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem J, 468(2): 293-301. 10.1042/BJ20141554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ, 2014. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol, 15(1): 7-18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Polato F, Callen E, Wong N, et al. , 2014. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J Exp Med, 211(6): 1027-1036. 10.1084/jem.20131939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, et al. , 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol, 153(3): 613-620. 10.1083/jcb.153.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, et al. , 2000. P53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol, 151(7): 1381-1390. 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, Durocher D, 2019. Shieldin—the protector of DNA ends. EMBO Rep, 20(5): e47560 10.15252/embr.201847560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, et al. , 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell, 136(3): 420-434. 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Tomida J, Takata KI, Bhetawal S, et al. , 2018. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J, 37(12): e99543 10.15252/embj.201899543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, 2009. Reaching out for the other end with p53-binding protein 1. Trends Biochem Sci, 34(5): 226-229. 10.1016/j.tibs.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ, 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA, 104(52): 20759-20763. 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Aroumougame A, Lobrich M, et al. , 2014. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev, 28(24): 2693-2698. 10.1101/gad.252478.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Yuan ZL, Cui YQ, et al. , 2018. Molecular basis for the inhibition of the methyl-lysine binding function of 53BP1 by TIRR. Nat Commun, 9: 2689 10.1038/s41467-018-05174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Takenaka K, Takeda S, 2010. PTIP promotes DNA double-strand break repair through homologous recombination. Genes Cells, 15(3): 243-254. 10.1111/j.1365-2443.2009.01379.x [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, et al. , 2004. 53BP1 is required for class switch recombination. J Cell Biol, 165(4): 459-464. 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GT, Chapman JR, Brandsma I, et al. , 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature, 521(7553): 541-544. 10.1038/nature14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Chini CCS, He M, et al. , 2003. The BRCT domain is a phospho-protein binding domain. Science, 302(5645): 639-642. 10.1126/science.1088753 [DOI] [PubMed] [Google Scholar]
- Zhang AL, Peng B, Huang P, et al. , 2017. The p53-binding protein 1-Tudor-interacting repair regulator complex participates in the DNA damage response. J Biol Chem, 292(16): 6461-6467. 10.1074/jbc.M117.777474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lou LH, Peng B, et al. , 2020. Nudix hydrolase NUDT16 regulates 53BP1 protein by reversing 53BP1 ADP-ribosylation. Cancer Res, 80(5): 999-1010. 10.1158/0008-5472.CAN-19-2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, et al. , 2013. 53BP1 regulates DSB repair using Rif1 to control 5' end resection. Science, 339(6120): 700-704. 10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]