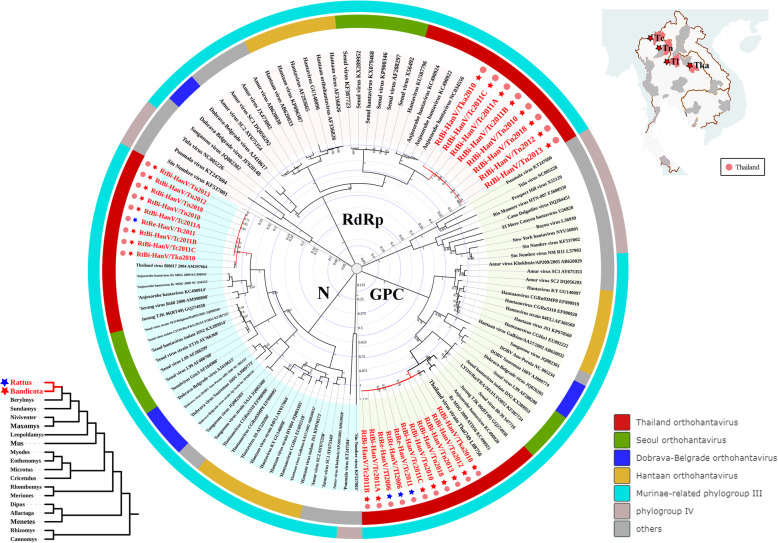
Fig. 3.
Phylogenetic trees based on the complete M segment-encoded glycoprotein precursor (GPC), L segment-encoded RdRp (RdRp), and S segment-encoded nucleocapsid protein (N) amino acid sequences of HanVs. Phylogenetic trees were constructed by the maximum likelihood method using the best-fit models (LG + G for GPC protein, LG + G + I for RdRp protein, and GTR + G + I for N protein). All HanVs found in this study are labeled in red. Host genus and location of each virus are labeled by the 5-point stars and dots of different colors. The outer color rings represent additional taxonomic information about these viruses. DOBV, Dobrava-Belgrade virus