Figure 1.
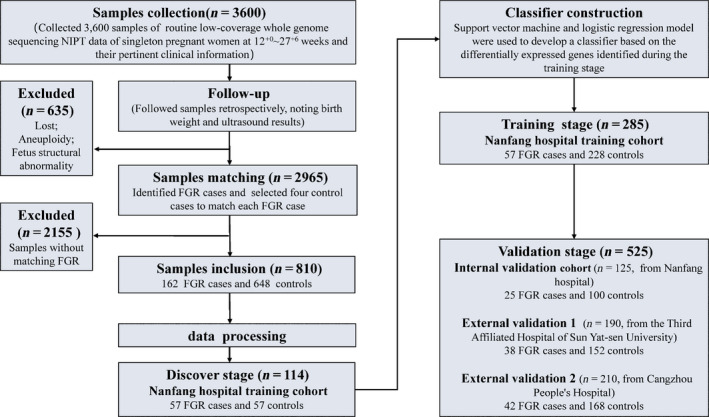
Flowchart of FGR classifier development based on low‐coverage whole‐genome sequencing. From three independent Chinese institutions, we collected 3600 samples of routine low‐coverage whole‐genome sequencing NIPT data from singleton pregnant women at 12+0–27+6 weeks, in addition to general clinical information. We used data from retrospective patient follow‐up reports, including pregnancy outcomes and final birthweight, to identify FGR cases and controls. According to gestational age at the time of maternal plasma sample extraction and fetal gender, four control cases were randomly selected to match each FGR case. Ultimately, we included 810 samples (162 FGR and 648 controls), which were divided into a Nanfang Hospital training cohort (285 samples from Nanfang Hospital), an internal validation cohort (125 samples from Nanfang Hospital), an external validation cohort 1 (190 samples from the Third Affiliated Hospital of Sun Yat‐sen University) and external validation cohort 2 (210 samples from Cangzhou People’s Hospital). In the discovery stage, gene promoters with a nucleosome footprint that differed between 57 FGR cases and 57 controls from Nanfang Hospital training cohort were identified. In the training stage, classifiers were developed via support vector machine (SVM) and logistic regression (LR) models, based on the genes with differentially coverages between the 57 FGR cases and 228 controls. In the validation stage, the optimal classifiers were further validated using the three validation cohorts.
