Figure 2.
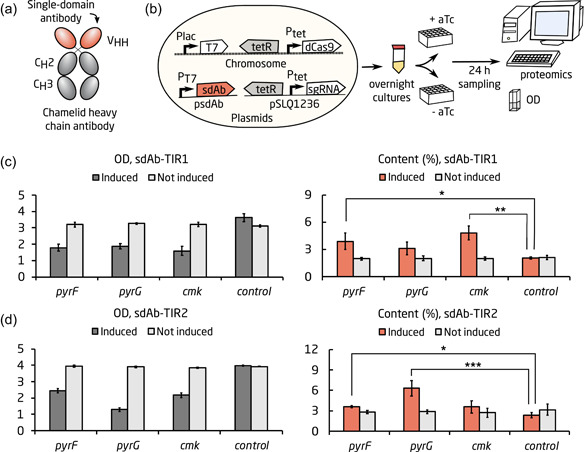
sdAb production in deep well plate. Application of the top‐performing growth switches pyrF, pyrG, and cmk for production of two expression‐optimized sdAbs with different translation initiation regions (TIR). (a) sdAbs are derived from the heavy chain of an antibody. (b) Experimental overview. (c) Growth and sdAb production after 24 hr for strains harboring psdAb‐TIR1 and sgRNA plasmids targeting pyrF, pyrG, and cmk. (d) Growth and sdAb production after 24 hr for strains harboring psdAb‐TIR2 and sgRNA plasmids targeting pyrF, pyrG, and cmk. For (c) and (d), the first bar graph shows OD and the second bar graph shows percent sdAb content. Cultures, where the CRISPRi system was induced, are shown in the dark gray (OD) or red (sdAb content). Uninduced cultures are shown in bright gray. The values were calculated as an average of three biological replicates. Error bars represent standard deviation of the replicates. A two‐tailed t test was used to check for significant difference between the strains; *p < .05; **p < .001; ***p < .0001. CRISPRi, CRISPR interference; OD, optical density; sdAb, single‐domain antibody [Color figure can be viewed at wileyonlinelibrary.com]
