Figure 2. Novel genes and gene models.
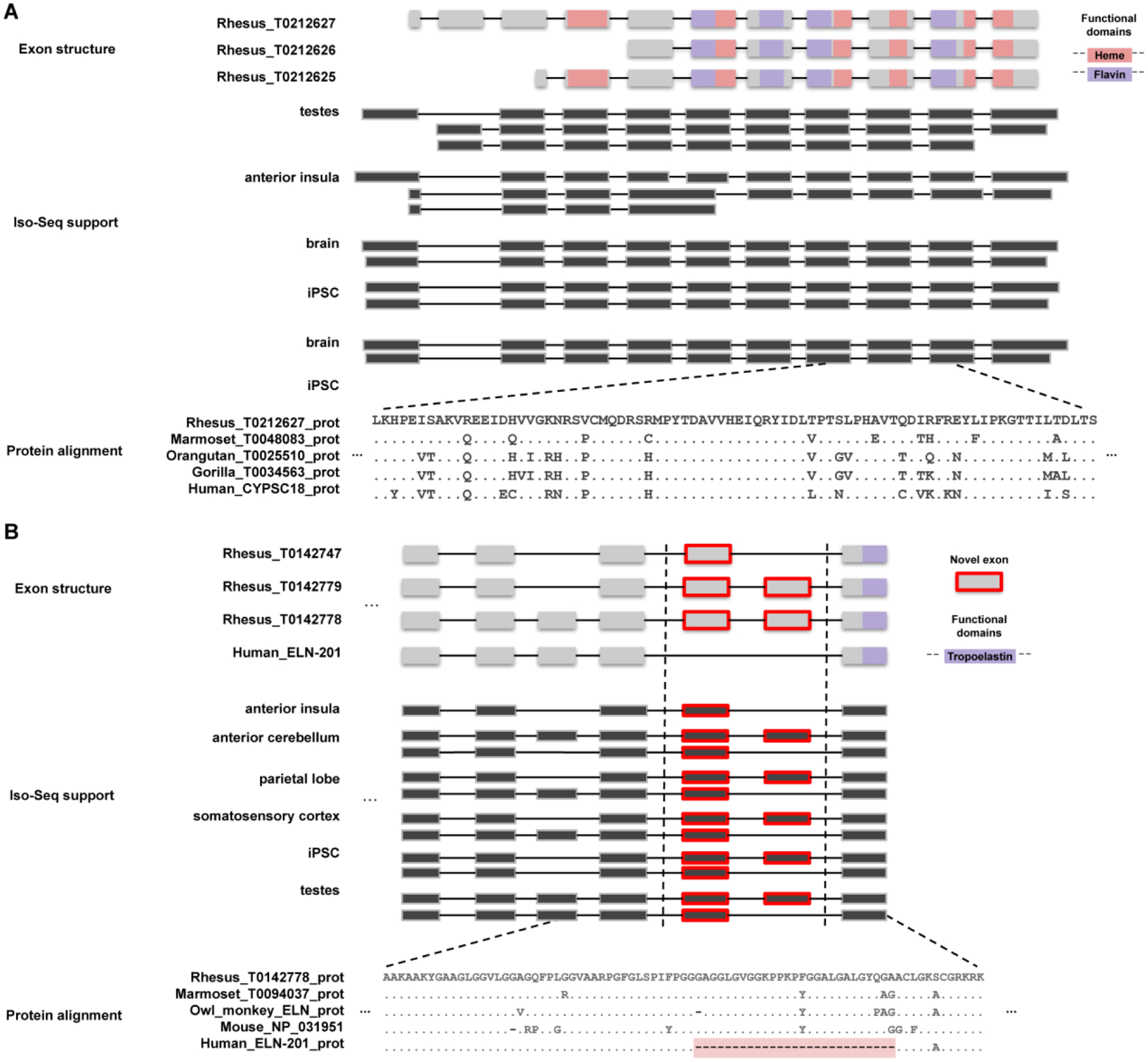
(A) A novel gene model with homology to the cytochrome p450 protein family is predicted by the AugustusPB mode of the CAT. The gene structure and protein domain architecture of three isoforms are shown (top). The predictions arose from supporting Iso-Seq reads from five tissues (middle). Orthologous novel genes are also predicted in marmoset, orangutan, and gorilla assemblies; a protein alignment (bottom) of those genes along with a human CYP2C18 protein is shown. (B) Two macaque isoforms in ELN (tropoelastin) are predicted by the AugustusPB mode of CAT and are supported by macaque Iso-Seq data but differ significantly from human by two exons. The gene structure and functional domains for the last seven exons of this gene are shown (top), along with a comparison to a human transcript model. These two protein-encoding exons are also observed in marmoset, owl monkey, and mouse, but not in apes, as a result of an ape-specific deletion (bottom) that changed the gene structure of tropoelastin.
