Figure 3. Macaque ZNF669 gene family expansion.
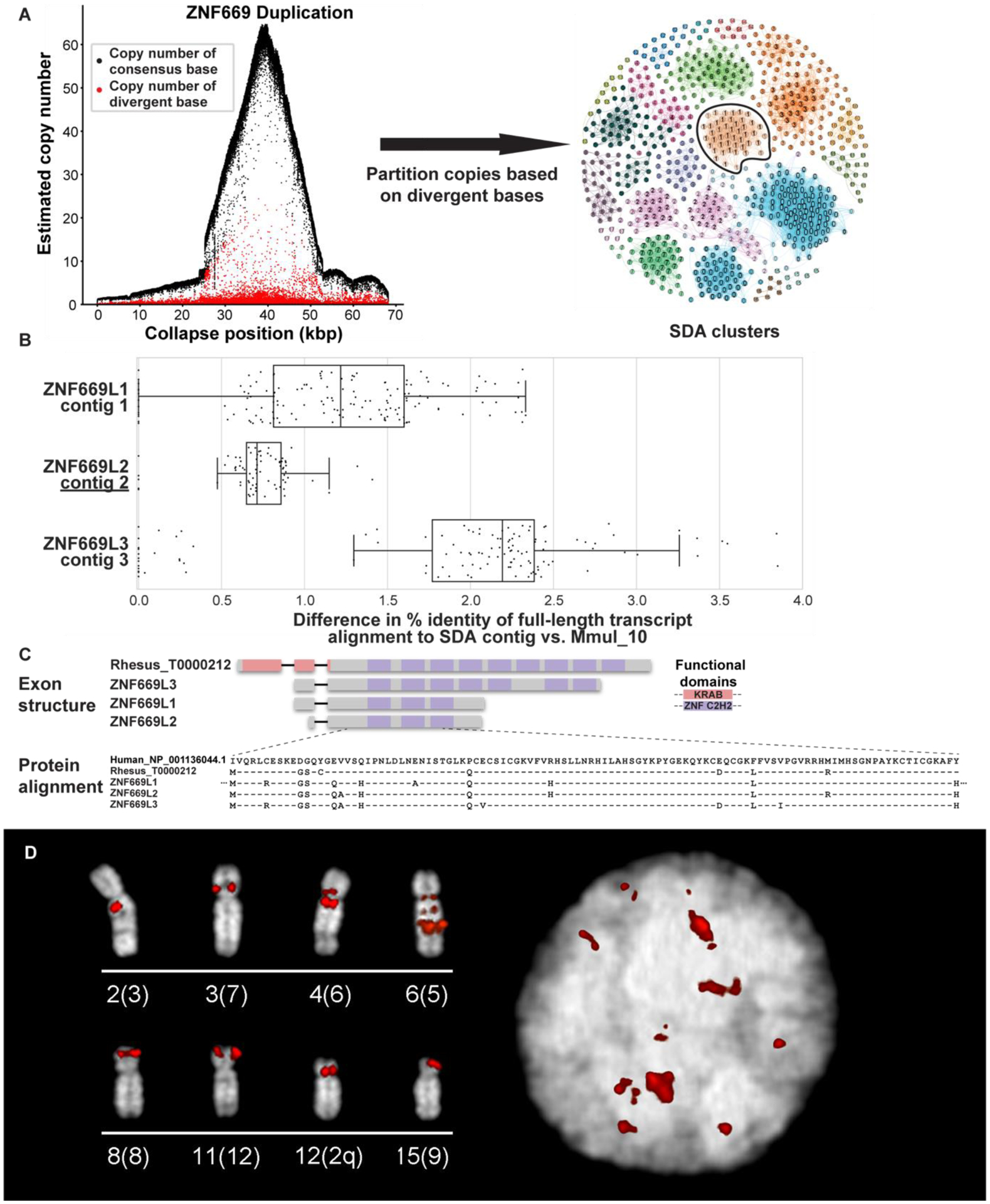
(A) A 68 kbp region of collapsed assembly corresponding to the ZNF669 gene family as indicated by the excess read depth and increased number of paralogous sequence variants (PSVs, red dots) that are diverged when compared to the consensus sequence (black dots). The highly identical copies were, thus, unresolved in Mmul_10 and predicted to be present in about 50 copies in macaque (left). Segmental Duplication Assembler (SDA) partitions the long reads into 19 distinct paralog clusters (colored and numbered) based on shared PSVs and assembled these clusters into 18 contigs. Vertices reflect individual PSVs and edges represent long-read sequences that contain both of the connected PSVs (right). SDA partitioned and assembled the remaining ZNF669 collapses into 35 additional contigs. The outlined PSV cluster corresponds to contig 2 in panel B. (B) Mapping of FLNC transcripts shows they align better to SDA-resolved contigs than the original assembly. (C) Annotation of these genes shows that these three contigs encode a highly expanded ZNF669 gene family where there is FLNC data supporting complete open reading frames that differ by only a few amino acids. (D) FISH with BAC CH250–540H16 as a probe corresponding to a ZNF669 locus demonstrate interchromosomal duplications (red) on interphase nucleus (right) and metaphase chromosomes (left), labeled by chromosome (human syntenic chromosome in parentheses).
