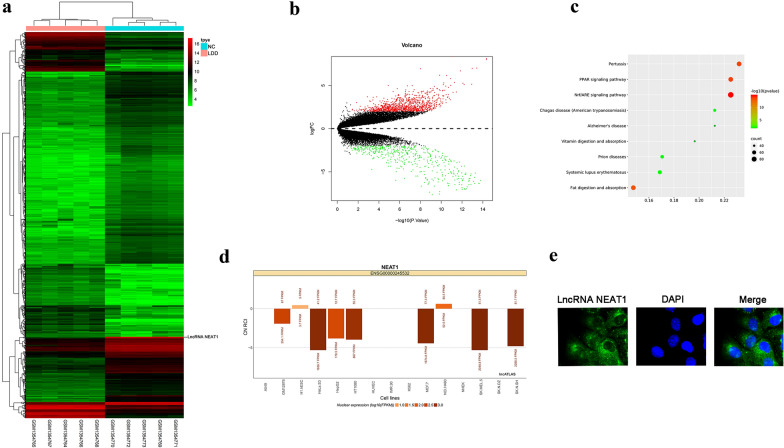
Fig. 1.
Bioinformatic analysis of GSE56081. a Heatmap of differentially expressed lncRNAs associated with IVD, red = high expression, green = low expression. b Volcano plot of differentially expressed lncRNAs associated with IVD, green dot represents down-regulated lncRNAs and red dot represents up-regulated lncRNAs in IVD tissue; black dot represents normally expressed lncRNAs. c KEGG pathway analysis for differentially expressed lncRNAs. d The abscissa represents gene ratio, and the ordinate represents enriched terms, the node color showed degree of enrichment ranging from low (green) to high (red), while the node size represented the frequency of the proteins in each enriched pathway group. e Subcellular localization plots of lncRNA NEAT1 displayed by lncATLAS. f Representative image of RNA FISH to confirm lncRNA NEAT1 location in nucleus pulposus cells (bars, 40 μm)