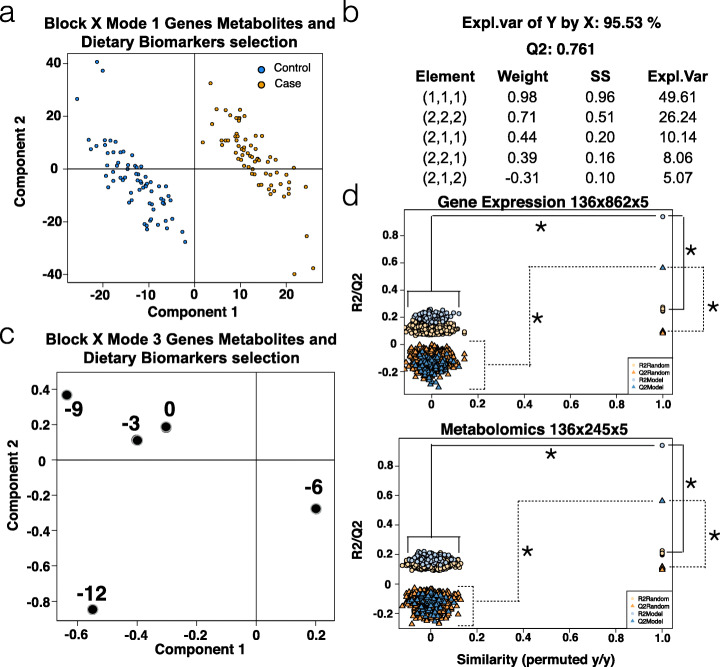
Fig. 2.
NPLS-DA analysis of TEDDY data. a NPLS-DA model mode 1 projection with combined gene expression, metabolites, and DBs showing separation between cases and controls. b R2, Q2, and element values for the NPLS-DA model. Each element is a triad containing the combination of mode components that captures the indicated explained variance. c NPLS-DA model mode 3 projection showing the relevance of early time points in the model as they have high absolute values. d VIP variable selection-validation by permutation analysis. For both gene expression and metabolomics datasets, the selected VIP-based features have significantly higher R2 (circles) and Q2 (triangles) values compared to a randomly selected set of features, (*) p < 0.001